1. Introduction
Gene delivery is a complex procedure where several obstacles must be overcome to successfully reach the target cell. Moreover, even after reaching the target cell, the biological effect is dependent on the effective delivery to the nucleus, for gene expression. Plasmid DNA (pDNA) has been considered a safer carrier for genes, but the pDNA itself must also be properly protected [
1]. An ideal pDNA carrier should promote good cargo protection, excellent colloidal stability, a high cellular uptake, an efficient endosomal escape and finally, an effective nucleus import and DNA unpacking. Hence, it is extremely important to develop delivery systems that can reunite all these characteristics being also able to target the cell/tissue of interest, for instance by using a hydrophilic protective corona for minimizing the non-specific interactions [
2].
Different approaches have been exploited for gene delivery. Naked DNA could be directly injected, but this approach would be limited to tissues reachable by direct injections (like skin or muscle) and also, it is unsuitable for systemic delivery due to the presence of serum nuclease [
3], which would immediately degrade the therapeutic gene. On the other hand, viral vectors could be applied for transferring the genetic material into the host cells. These vectors are highly effective on both gene delivery and expression. However, the use of viral vectors has some drawbacks, as the possibility to provoke immune responses, the associated costs, the difficulty for the preparation, and the limit to just carry small amounts of genetic material [
4].
Non-viral vectors have stood as safer alternatives to be applied for gene transfer. For the delivery of pDNA, systems are usually composed of cationic polymers or lipids with the ability to interact with the negatively charged DNA through electrostatic interactions leading to polyplexes and lipoplexes formation [
5]. Non-viral vectors are usually safe (causing a low immune response), easily prepared, have a low production cost, and can be easily produced on large scale. Another important characteristic of these vectors is the ability to transfer different and large transgenes, being also able to be stored for long periods due to their stability [
6].
Meanwhile, systemic delivery is a real challenge for these non-viral vectors since they need to survive in the bloodstream without being degraded or captured by cellular defence mechanisms. Also, when reaching the target tissue, the systems must go across the tissue and bind specifically to the target cells. After this internalization process, it is further required to surpass intracellular barriers such as the endosomal escape, the cytoplasm traffic, and finally, enter the nucleus [
7]. So, the ability of non-viral vectors to overcome these barriers will dictate their efficiency.
To develop pDNA-nanocarriers, different polymers have been applied. Linear or branched polyethyleneimine (PEI) has been extensively studied in a wide range of molecular weights, to promote the pDNA in vitro delivery [
8,
9]. However, some concerns regarding the high toxic behaviour of high molecular weight PEI (> 25 kDa) have limited its application. On the other hand, low molecular weight PEI demonstrated to be less toxic but showed poor transfection activity [
10]. On the other hand, chitosan (CH), a natural cationic polymer that is a linear polysaccharide, has also been extensively used for several biomedical and pharmaceutical applications due to the advantageous properties, like biocompatibility and muco-adhesivity [
11,
12]. For instance, in 2011, Gaspar and co-workers developed a chitosan-based carrier to deliver the p53 encoding pDNA. With the use of this p53-CH delivery system, the authors were able to reinstate the levels of the P53 protein in cancer cell lines [
12]. However, the CH polymer can also present some limitations regarding its application in gene delivery, due to the poor solubility in physiological conditions (pKa value around 6.3–6.4), which reinforces the need to proceed optimizing these systems.
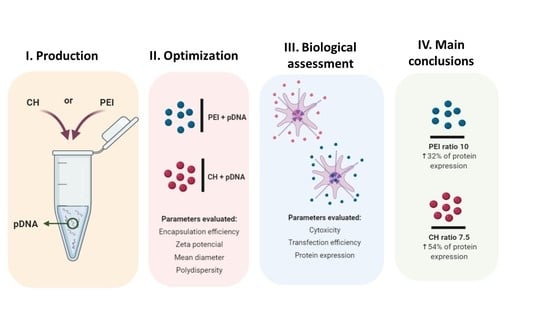
Overall, in this research work, a comparative study was carried out to develop efficient pDNA nanocarriers using complexation between pDNA with different sizes and CH or PEI as cationic polymers. Therefore, the parameters for complexation between the selected polymers and pDNA were optimized, to find the most suitable conditions that led to improved systems, in terms of encapsulation efficiency, zeta potential, and size. Besides, this research work will enable the readers to access a full range of conditions that could be applied for both PEI and CH polyplexes formulation as well as will enable them to predict the kind of properties concerning the encapsulation efficiency, zeta potential, mean diameter, polydispersity, cytotoxicity, cellular uptake and protein expression that the systems can provide, considering the size of the target pDNA. Afterwards, the best polyplexes were characterized in a cellular environment, where a therapeutic nano-formulation based on a pDNA with a therapeutic target (in this case the p53 encoding pDNA) was used to investigate the ability to promote P53 protein expression.
3. Results and Discussion
3.1. Plasmid DNA Loading Efficiency in Polymer Nanoparticles
The encapsulation efficiency is one of the parameters that must be evaluated when it is intended for the selection and optimization of delivery systems designed to encapsulate, transport and release drugs or biomolecules. An ideal nanocarrier should be able to transport the highest amount of the target biomolecule possible, enabling its protection and delivery.
In this study, to produce nanocarriers, the pDNA/polycation complexes were formed by electrostatic interactions between the negatively charged pDNA phosphate groups and protonated nitrogen atoms of the chosen polycations, as previously mentioned [
16]. The work started with a screening of different pDNAs:CH/PEI ratios to evaluate which one could lead to higher pDNA encapsulation. The plasmids under study were pVAX-GFP, pcDNA3-FLAG-p53, and p1321 HPV-16 E6/E7 with the respective sizes of 3, 6, and 9 kbp. Through this screening, it was verified that the CH-based polyplexes presented higher encapsulation efficiencies for N/P ratios of 1 and 7.5 with the largest and smallest pDNA (
Figure 1). It was also verified a tendency for an increased EE with the increase on N/P ratios until the highest EE value was reached (near 100% to pVAX-GFP and pcDNA3-FLAG-p53 and 90% to p1321 HPV-16 E6/E7), and then a sharp decrease of the EE was achieved for all the pDNAs. The lower EE were obtained for lower and higher polymer concentrations. In the case of higher ratios, the values obtained could be due to an excessive high positive charge causing a limitation on DNA access and interaction. Similarly, Gan and Wang (2007) have demonstrated that increasing the chitosan concentrations could lead to an increase in the solution viscosity which could be a major contributor to the decrease of the EE. Furthermore, the high viscosity associated with increased chitosan concentration hinders EE by deterring the biomolecules movement around chitosan molecular chain [
17]. Concerning the low EE levels observed for the lower polymer ratios, it is suggested that the polymer was insufficient to induce the formation of polyplexes, due to the very low amount of available chitosan, limiting the interaction and encapsulation of pDNA.
Regarding the EE achieved for the PEI-based polyplexes, it was clear that for all the pDNA in the study, the best N/P ratios were above 1, and the maximum EE (very close to 100%) remained constant for all the higher N/P ratios (
Figure 1). This result agrees with several studies that show the outstanding EE of PEI and its derivatives, regardless of the processing conditions. Despite their bio-applications, the high toxicity of PEI limits the efficiency of gene transfer in vitro and especially
in vivo [
15]. To fight this, several approaches have been reported to increase the gene transfection efficiency of PEI-based gene carriers while decreasing their intrinsic cytotoxicity [
18].
3.2. Zeta Potential and Hydrodynamic Size Measurements
The positive surface charge is a very important requirement to be considered for any carrier to be used as an efficient gene delivery system. Moreover, the entry in the intracellular compartment will be simplified if the delivery system presents a small size (<200 nm) [
16,
17]. Thus, the zeta potential and size of the pDNA-loaded polyplexes were evaluated. The values obtained from the zeta potential measurements represent the value of the electrostatic potential at the plane of shear and, zeta potential values near ±30 mV are representative of stabilized particles [
19].
Figure 2 shows the zeta potential values obtained for each pDNA conjugated with CH or PEI. Through the analysis of these results, it was possible to observe that for all the polyplexes, the zeta potential presented negative values up until the N/P ratio of 1. Meanwhile, the higher surface charge for CH-based polyplexes is comprised, in general, between N/P ratio 2.5 and 10 and, among these N/P ratios the desirable +30 mV was also accomplished. The highest value achieved was above 35 mV at N/P ratio 2.5 for the p53 encoding pDNA, and then the zeta potential decreased. It was also observed that for the pDNA with higher molecular weight (p1321 HPV-16 E6/E7), the decrease in the zeta potential was more evidenced for ratios above 7.5, while, for the smaller pDNA, a more pronounced difference was achieved only for N/P ratios higher than 15. For higher N/P ratios, the surface charge of the CH-based polyplexes suffered a slight decrease and then a stabilization. This decrease could hypothetically be related to a possible rearrangement in the polyplex structure, enabling a higher presence of the pDNA at the surface of the particles.
For PEI-based polyplexes, the highest zeta potential measurements were observed for higher N/P ratios when compared with CH-based polyplexes, being the values obtained very close to +30 mV. When the PEI values are closely analysed, it is observed a relationship between the ratios where it was found the stabilization of the EE and the maximum zeta potential. The zeta potential can also be used to determine whether a charged active material is encapsulated within the centre of the polyplex or on the surface. Thus, and considering the results obtained, it is suggested that the polyplexes produced present a “shell” made essentially of polymer being the genetic information fully protected inside the carriers. These polyplexes also presented a decrease in their zeta potential, but only for the highest N/P ratio used (N/P ratio of 50). This behaviour was previously described for aqueous solutions of two oppositely charged polyelectrolytes. In 2005, Zhang and Shklovskii showed that an increase in the polycation concentration ratio increases the size of the complexes reaching a maximum at the isoelectric point, and then decreases again accompanied by a “charge reversal” phenomenon [
19,
20]. To follow the characterization of the polyplexes, and considering the results achieved for the zeta potential and EE, the assessment of the hydrodynamic size was only performed for the best three ratios. Thus, for CH-based polyplexes, the N/P ratios of 5, 7.5, and 10 were selected, while the PEI-based polyplexes characterized were for the N/P ratios of 7.5, 10, and 15.
As shown in
Figure 3, the size of CH-based polyplexes decreased with the increase of the polymer ratio for pVAX-GFP and p53 encoding pDNA, while for p1321 HPV-16 E6/E7 the size decreased between N/P ratio 5 and 7.5, but then stabilized between N/P ratio 7.5 to 10. In the case of PEI-based polyplexes, de hydrodynamic size decreased between N/P ratio 7.5 to 10 and then stabilized between N/P ratio 10 to 15, for all the studied pDNA, but with a more pronounced effect for pVAX-GFP. Overall, it was found that the smaller nanocarriers were obtained at N/P ratio 15 and 10 for PEI and 7.5 and 10 for CH. Moreover, the increase of polydispersity, namely in the PEI-based polyplexes, could be induced by interactions through the accessible unbound DNA chains in one polyplex with the unbound PEI of another polyplex, resulting in some aggregation. On the other hand, polydispersity at relatively low ratios (as in the case of CH-based polyplexes) could be related to the initially formed polyplexes since they tend to agglomerate due to the nearly neutral surface charge of the polyplexes [
21].
Overall, the PEI-based polyplexes showed to be smaller than the CH-based polyplexes, however, for the studied conditions, PEI polymer presented a very high PDI when compared with CH.
As mentioned above, the entry of large particles into the cell it is not easy, and their structure is not stable enough to resist the low pH environment and lysosomal enzymes [
22]. Thus, the complete characterization and selection of suitable nanocarriers must include the evaluation of the biological response of the cells transfected with the polyplexes produced. In this case, the biological effect was assessed for the CH- and PEI-based polyplexes that presented the best values in terms of EE, Zeta potential, and hydrodynamic size. These conditions led to the selection of the N/P ratio of 7.5 for CH-based polyplexes and N/P ratio of 10 for PEI-based polyplexes.
3.3. Cytotoxicity Evaluation
In vitro studies using cancer and non-cancer cell lines to predict human response, typically play a vital role in acquiring relevant information about the behaviour of these systems in a biological environment before further
in vivo application of the formulations [
23,
24]. In this context, the toxicity levels of the formulations studied (PEI and CH nanoparticles) were accessed. The influence of the different polyplexes in HeLa cancer cells and human dermal fibroblasts (hFib) was studied and compared to search for the possible intracellular toxicity often associated with these cationic nanocarriers-mediated uptake. The results obtained (
Figure 4) revealed that CH-based polyplexes loaded with different pDNA did not induce cytotoxicity, as the slight differences are not statistically significant in comparison to the negative controls. This was a predictable result since previous studies using pDNA-CH nanocomplexes have demonstrated low cytotoxicity for similar systems [
12]. Therefore, this polymer has also been shown to destabilize the lipid bilayer, thus facilitating its cellular uptake which could be extremely advantageous for the efficient delivery of the desired genetic information [
25].
Concerning the PEI-based polyplexes, statistical relevance was found for the cytotoxicity induced by p53-encoding pDNA-loaded polyplexes and the negative control but only for the hFib cells. This result could demonstrate potential
in vivo toxicity of these carriers for non-tumour cells which could be a major concern in its future application [
26]. This cytotoxicity presented by the PEI-based polyplexes could be explained by the higher polymer ratio applied. In fact, in literature, it was demonstrated that cationic polymers can induce cytotoxic effects at high concentrations because of their strong electrostatic interactions with the cell membrane, which can lead to destabilization and eventually rupture of the cell [
27]. Also, some previous works made attempts to obtain information about the composition of the PEI-based polyplexes, demonstrating that large amounts of the PEI remain free and not involved in complex formation. These free PEI molecules presented higher toxicity than PEI bound as the polyplex [
28]. Overall, these results seem to suggest that CH-based systems could be more promising for safer pDNA delivery than PEI-based formulations.
3.4. Evaluation of the Transfection Efficiency
Previous studies regarding the uptake mechanisms for CH or PEI nanoparticles indicated that CH-based polyplexes are mainly uptake through the clathrin-based endocytic mechanism, while PEI-based polyplexes have presented preference for the caveolar pathway in HeLa cells [
29,
30,
31]. As referred, it is commonly accepted that complexes produced between DNA/cationic polymers are taken up by cells via endocytosis, however, further stages of their endosomal release, namely the transport along with the cytoplasm and also, the transfer to the nucleus and further DNA release, are less well understood. One of the mechanisms of the endosomal release of the complexes into the cytoplasm is based on the proton sponge hypothesis [
32]. Research studies have demonstrated that several cationic polymers, such as PEI and CH, can buffer endosomal acidification causing an accumulation of protons which promotes an influx of chloride anions. This process increases osmotic pressure which promotes entry of water, and thereby the disruption of the endosome. It is worth mentioning that membrane destabilization by free polycation has been proposed as another mechanism that could also contribute to the endosomal release of the highly charged polymers [
33].
Concerning this, the DNA delivery in eukaryotic cells is a multistep process that begins with the condensation of DNA, the introduction of DNA into the systemic circulation, and targeted delivery to specific cells followed by cellular uptake, endosomal release, nuclear transport, and unpacking of the carrier/DNA polyplexes before the final step of translation in eukaryotic cells [
3]. In the present work, to explore the DNA delivery process, the behaviour of these molecules was monitored by confocal laser scanning microscopy. The cell live imaging after 1 h of transfection (
Figure 5) showed the entry into the cell for all the pDNA/cationic polymers studied. The carriers of smaller pDNA promoted faster cell transfection than the ones delivering the largest pDNA, for both CH- and PEI-based polyplexes. The system that seems to be faster for transfection was the CH with pVAX-GFP. In this case, cells presented a large number of labelled-particles inside the nucleus, which can be indicative of the efficiency of these systems entering the nucleus. Through these results, it is predictable that CH-based polyplexes could be more efficient on transfection when compared with the PEI-based polyplexes, which is also following the previous cytotoxicity results.
Literature mentioned that lower transfection levels at higher polymer ratios may be induced by competition between the excess of cationic polymer present in the formulation. This extra polymer can bind to the cell surface, preventing complexes from being efficiently internalized [
27]. This explanation about the excess of the polymer could be the reason for PEI nanoparticles present a slightly higher cytotoxic behaviour as well as a lower transfection rate since for this case a higher ratio is applied when compared with the CH ratio chosen. Another reasonable explanation could be a slight difference in the zeta potential presented for both carriers. The CH-based polyplexes presented the highest mV value and this could also favour the interaction with the negative cell membrane.
Concerning the largest pDNA studied, the behaviour for CH- and PEI-based polyplexes was very similar, since in both cases cells presented a low transfection rate and when transfected, few labelled-particles were found in the nucleus, which can suggest that low protein expression could be achieved. Different studies have already been performed to compare the influence of the pDNA size on the transfection effectiveness, being verified that actually, as short as the pDNA is, as higher is the transfection rate. Kreiss and collaborators (1999) hypothesized, based on the scientific evidence from their research work, that the nuclear entry via the nuclear pores might be dependent on plasmid size (as shorten was the pDNA as effective was the gene transfer process) [
34].
3.5. P53 Protein Expression
The formation of these polymeric nanoparticles occurs by the entrapment of the genetic material into the polymer matrix and thus, the release rate of pDNA is dependent on cationic polymer biodegradation. Several studies have suggested that DNA unpacking is one of the major intracellular barriers to effective expression [
35]. It has been recognized that a balance between DNA protection and its ability to dissociate from the nanoparticles must be achieved to obtain efficient protein expression [
27].
To evaluate the real ability of these polyplexes to transfer genetic information across the cell to reach the nucleus, HeLa cells were transfected with both polyplexes conjugated with the p53 encoding pDNA, and then P53 protein levels were measured through an ELISA analysis. The cell line was previously used to assess the expression of P53 after a transfection experiment and showed to be very sensitive to the p53 encoding gene treatment [
33,
36]. From the analysis of
Figure 6, it is possible to observe that, although the chosen cells already present a basal expression of the P53 protein, when transfected with CH-p53 encoding pDNA polyplexes, this level increases around 54.2%, and when transfected with PEI-loaded polyplexes with the same pDNA, the protein level increases around 31.96%, in comparison with the P53 basal levels.
As mentioned, the free polycation in the DNA/polymer nanoparticles dispersion has shown to be mandatory to an efficient transfection, since it helps on the endosomal scape. Some studies have shown that DNA/polycation complexes alone cannot trigger their endosomal escape through the proton sponge mechanism without a sufficiently high content of free polycation inside the endosome. When nanoparticles find the mechanism to escape from the endosome, the DNA released from the polycation migrates into the nucleus for further transcription. However, it was also described that gene expression decreases when DNA was either tightly or loosely bound to the polycations, such as PEI or CH [
35,
37]. The tight bound and highly stable polyplexes will be readily endocytosed but possibly not disassembled to access the transcription machinery. On the other hand, DNA weakly bound to the polycation will produce complexes that will dissociate prematurely in the medium and not even be endocytosed by cells [
38].
Regarding our results from confocal microscopy analysis, it was possible to observe a slight increase of the p53 nanoparticles entrapment for CH-based polyplexes, a behaviour that could suggest a lower P53 protein expression for the PEI-based polyplexes. Besides this, we can also suggest that the decreased protein expression could be related to a weak linkage of DNA with PEI, when compared with the CH/DNA polyplexes, which could lead to rapid degradation of the delivered gene. This degradation process decreases the amount of viable genes to be used for the transcription machinery, which will also decrease the amount of the final P53 protein produced.
It is also important to refer that using the DNA polymer complexation method, we were able to accomplish better P53 protein expression using the CH-based polyplexes than Gaspar and co-workers (2011). In that previous study, an ionotropic gelation technique was performed between CH and TPP (used as polyanion) and where the p53 encoding pDNA vector was also added. To accomplish the production of the nanocapsules, TPP+pDNA was dropwise added to the chitosan solution. However, using this method the authors only accomplish 40% of P53 expression [
12].
Concerning all the above mentioned, an effective gene delivery nano complex should provide an appropriate balance between the DNA and polycation, to promote not only binding strength and stability but also, guarantee that DNA can dissociate intracellularly for gene expression.
4. Conclusions
To promote gene transfer, non-viral systems such as nanoparticles have arisen as efficient and safe delivery tools. Several cationic polymers have been used in the nanocarriers development, and in this work, different pDNA were conjugated with CH and PEI to search for the suitable nano complex combination. Regarding the surface charge and encapsulation efficiency characterization, it was verified that, depending on the ratios used, it could be possible to define suitable conditions to prepare positively charged nanocarriers highly efficient on DNA loading (around 80 to 100% for the best conditions), which is crucial for the application. Additionally, the biological effect was assessed for the CH- and PEI-based polyplexes that presented the best properties in terms of EE, Zeta potential and hydrodynamic size (N/P ratio 7.5 for CH and N/P ratio 10 for PEI). Through the cytotoxic profile, it was observed biocompatibility for CH-based polyplexes, however, for p53-encoding pDNA/PEI polyplexes it was verified slight toxicity in normal cells which could be a handicap for future therapeutic application of these polyplexes. Regarding the transfection efficiency, it was verified that all the pDNA /cationic polymers systems studied were able to transfect the cells, but a higher efficiency was reached for the smaller pDNA. Finally, the ability of the polyplexes to promote P53 protein expression was also addressed using HeLa cancer cells. From the results obtained, the P53 levels increased around 54.2% and 32% when CH- and PEI-based polyplexes were respectively applied.
Overall, it is possible to refer that an effective gene delivery nano complex should provide an appropriate balance between the DNA and the polycation. The chosen conditions for the polyplexes formulation should consider parameters such as EE, size, zeta potential and polydispersity being also needed to perform a careful polymer selection. Besides, given the presented results and the used complexation conditions it was observed that the polyplexes using an N/P ratio 7.5 of CH presented less cytotoxicity, were more efficient regarding cell uptake and finally led to an increased protein expression when compared with the PEI polyplexes, which suggests that CH polyplexes can be more suitable for pDNA delivery.