Molecularly Imprinted Ligand-Free Nanogels for Recognizing Bee Venom-Originated Phospholipase A2 Enzyme
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
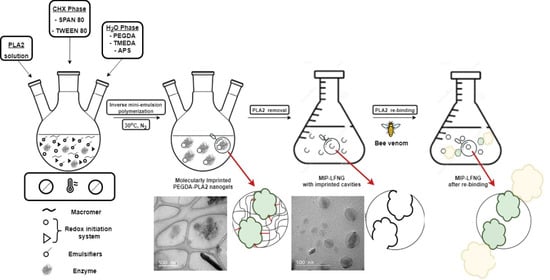
2.2. Synthesis and Purification of LFNGs for Recognizing and Retaining Bee Venom-Originated PLA2
2.3. Characterization Methods and Instruments
2.3.1. Structural and Morphological Characterization of LFNGs
2.3.2. Batch Binding Experiments Assisted by Activity Measurements of PLA2
2.3.3. Cytotoxicity Study of LFNGs
3. Results
3.1. Synthesis of LFNGs
3.2. Structural and Morphological Characterization of LFNGs
3.2.1. FT-IR Spectroscopy
3.2.2. TGA Investigation
3.2.3. DLS Investigation
3.2.4. TEM Images
3.3. Binding Properties of LFNGs
3.4. Cytotoxicity Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Freeman, T.M. Hypersensitivity to Hymenoptera Stings. N. Engl. J. Med. 2004, 351, 1978–1984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaloske, R.H.; Dennis, E.A. The phospholipase A2 superfamily and its group numbering system. Biochim. Biophys. Acta 2006, 1761, 1246–1259. [Google Scholar] [CrossRef] [PubMed]
- Cajal, Y.; Jain, M.K. Synergism between Mellitin and Phospholipase A2 from Bee Venom: Apparent Activation by Intervesicle Exchange of Phospholipids. Biochemistry 1997, 36, 3882–3893. [Google Scholar] [CrossRef] [PubMed]
- Tuĭchibaev, M.U.; Akhmedova, N.U.; Muksimov, F.A. Hemolytic effect of phospholipase A2 and orientotoxin from venom of the great hornet, Vespa orientalis. Biokhimiia 1988, 53, 434–443. [Google Scholar] [PubMed]
- Zambelli, V.O.; Picolo, G.; Fernandes, C.A.H.; Fontes, M.R.M.; Cury, Y. Secreted Phospholipases A2 from Animal Venoms in Pain and Analgesia. Toxins 2017, 9, 406. [Google Scholar] [CrossRef] [Green Version]
- Mingarro, I.; Pérez-Payá, E.; Pinilla, C.; Appel, J.R.; Houghten, R.A.; Blondelle, S.E. Activation of bee venom phospholipase A2 through a peptide-enzyme complex. FEBS Lett. 1995, 372, 131. [Google Scholar] [CrossRef] [Green Version]
- Almeida, R.A.M.B.; Olivo, T.E.T.; Mendes, R.P.; Barraviera, S.R.C.S.; Souza, L.R.; Martins, J.G.; Hashimoto, M.; Fabris, V.E.; Junior, R.S.F.; Barraviera, B. Africanized honeybee stings: How to treat them. Rev. Soc. Bras. Med. Trop. 2011, 44, 755–761. [Google Scholar] [CrossRef] [Green Version]
- Muraro, A.; Roberts, G.; Worm, M.; Bilò, M.B.; Brockow, K.; Rivas, M.F.; Santos, A.F.; Zolkipli, Z.Q.; Bellou, A.; Beyer, K.; et al. Anaphylaxis: Guidelines from the European Academy of Allergy and Clinical Immunology. Allergy 2014, 69, 1026–1045. [Google Scholar] [CrossRef]
- Schumacher, M.J.; Egen, N.B.; Tanner, D. Neutralization of bee venom lethality by immune serum antibodies. Am. J. Trop. Med. Hyg. 1996, 55, 197–201. [Google Scholar] [CrossRef]
- Santos, K.S.; Stephano, M.A.; Marcelino, J.R.; Ferreira, V.M.R.; Rocha, T.; Caricati, C.; Higashi, H.G.; Moro, A.M.; Kalil, J.E.; Malaspina, O.; et al. Production of the first effective hyperimmune equine serum antivenom against Africanized bees. PLoS ONE 2013, 8, e79971. [Google Scholar] [CrossRef]
- Laustsen, A.H.; Gutiérrez, J.M.; Knudsen, C.; Johansen, K.H.; Bermúdez-Méndez, E.; Cerni, F.A.; Jürgensen, J.A.; Ledsgaard, L.; Martos-Esteban, A.; Øhlenschlæger, M.; et al. Pros and cons of different ther-apeutic antibody formats for recombinant antivenom development. Toxicon 2018, 146, 51–75. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.G.A.; Corteling, R.L.; To, H.P.; Bhogal, G.; Landon, J. A novel Fab-based antivenom for the treatment of mass bee attacks. Am. J. Trop. Med. Hyg. 1999, 61, 361–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pucca, M.B.; Cerni, F.A.; Oliveira, I.S.; Jenkins, T.P.; Argemí, L.; Sørensen, C.V.; Ahmadi, S.; Barbosa, J.E.; Laustsen, A.H. Bee Updated: Current knowledge on bee venom and bee envenoming therapy. Front. Immunol. 2019, 10, 2090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pessenda, G.; Silva, L.C.; Campos, L.B.; Pacello, E.M.; Pucca, M.B.; Martinez, E.Z.; Barbosa, J.E. Human scFv antibodies (Afribumabs) against Africanized bee venom: Advances in melittin recognition. Toxicon 2016, 112, 59–67. [Google Scholar] [CrossRef]
- Jenkins, T.P.; Fryer, T.; Dehli, R.I.; Jürgensen, J.A.; Fuglsang-Madsen, A.; Føns, S.; Laustsen, A.H. Toxin Neutralization Using Alternative Binding Proteins. Toxins 2019, 11, 53. [Google Scholar] [CrossRef] [Green Version]
- Oh, J.K.; Drumright, R.; Siegwart, D.J.; Matyjaszewski, K. The development of microgels/nanogels for drug delivery applications. Prog. Polym. Sci. 2008, 33, 448–477. [Google Scholar] [CrossRef]
- Wani, T.U.; Rashid, M.; Kumar, M.; Chaudhary, S.; Kumar, P.; Mishra, N. Targeting aspects of nano-gels: An overview. Int. J. Pharm. Sci. Nanotechnol. 2014, 7, 2612–2630. [Google Scholar] [CrossRef]
- Patoo, T.S.; Khanday, F.; Qurashi, A. Prospectus of advanced nanomaterials for antiviral properties. Mater. Adv. 2022, 3, 2960–2970. [Google Scholar] [CrossRef]
- Anooj, E.S.; Charumathy, M.; Sharma, V.; Vibala, B.V.; Gopukumar, S.T.; Beema Jainab, S.I.; Vallinayagam, S. Nanogels: An overview of properties, biomedical applications, future research trends and developments. J. Mol. Struct. 2021, 1239, 130446. [Google Scholar] [CrossRef]
- Magadán, S.; Mikelez-Alonso, I.; Borrego, F.; González-Fernández, Á. Nanoparticles and trained immunity: Glimpse into the future. Adv. Drug Deliv. Rev. 2021, 175, 113821. [Google Scholar] [CrossRef]
- Whitcombe, M.J.; Kirsch, N.; Nicholls, I.A. Molecular imprinting science and technology: A survey of the literature for the years 2004–2011. J. Mol. Recognit. 2015, 27, 297–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ulusoy, M.; Aslıyüce, S.; Keskin, N.; Denizli, A. Beauvericin purification from fungal strain using molecularly imprinted cryogels. Process Biochem. 2022, 113, 185–193. [Google Scholar] [CrossRef]
- Deyev, S.M.; Lebedenko, E.N. Modern Technologies for Creating Synthetic Antibodies for Clinical application. Acta Nat. 2009, 1, 32–50. [Google Scholar] [CrossRef]
- Long, Y.; Li, Z.; Bi, Q.; Deng, C.; Chen, Z.; Bhattachayya, S.; Li, C. Novel polymeric nanoparticles targeting the lipopolysaccharides of Pseudomonas aeruginosa. Int. J. Pharm. 2016, 502, 232–241. [Google Scholar] [CrossRef]
- Xu, S.; He, H.; Liu, Z. New Promises of Advanced Molecular Recognition: Bioassays, Single Cell Analysis, Cancer Therapy, and Beyond. Chin. J. Chem. 2022, 40, 635–650. [Google Scholar] [CrossRef]
- Hoshino, Y.; Koide, H.; Furuya, K.; Haberaecker, W.W., III; Lee, S.H.; Kodama, T.; Kanazawa, H.; Oku, N.; Shea, K.J. The rational design of a synthetic polymer nanoparticle that neutralizes a toxic peptide in vivo. Proc. Natl. Acad. Sci. USA 2012, 109, 33–38. [Google Scholar] [CrossRef] [Green Version]
- Hoshino, Y.; Shea, K.J. The evolution of plastic antibodies. J. Mater. Chem. 2011, 21, 3517–3521. [Google Scholar] [CrossRef]
- Hoshino, Y.; Koide, H.; Urakami, T.; Kanazawa, H.; Kodama, T.; Oku, N.; Shea, K.J. Recognition, Neutralization, and Clearance of Target Peptides in the Bloodstream of Living Mice by Molecularly Imprinted Polymer Nanoparticles: A Plastic Antibody. J. Am. Chem. Soc. 2010, 132, 6644–6645. [Google Scholar] [CrossRef] [Green Version]
- Hoshino, Y.; Urakami, T.; Kodama, T.; Koide, H.; Oku, N.; Okahata, Y.; Shea, K.J. Design of Synthetic Polymer Nanoparticles that Capture and Neutralize a Toxic Peptide. Nano Micro Small 2009, 5, 1562–1568. [Google Scholar] [CrossRef] [Green Version]
- Hayakawa, N.; Yamada, T.; Kitayama, Y.; Takeuchi, T. Cellular Interaction Regulation by Protein Corona Control of Molecularly Imprinted Polymer Nanogels Using Intrinsic Proteins. ACS Appl. Polym. Mater. 2020, 2, 1465–1473. [Google Scholar] [CrossRef]
- Takeuchi, T.; Kitayama, Y.; Sasao, R.; Yamada, T.; Toh, K.; Matsumoto, Y.; Kataoka, K. Molecularly Imprinted Nanogels Acquire Stealth In Situ by Cloaking Themselves with Native Dysopsonic Proteins. Angew. Chem. Int. Ed. Engl. 2017, 56, 1–6. [Google Scholar] [CrossRef]
- Radu, A.L.; Gavrila, A.M.; Cursaru, B.; Spatarelu, C.P.; Sandu, T.; Sarbu, A.; Teodorescu, M.; Perrin, F.X.; Iordache, T.V.; Zaharia, A. Poly (ethylene Glycol) Diacrylate-Nanogels Synthesized by Mini-emulsion Polymerization. Mater. Plast. 2019, 56, 514–519. [Google Scholar] [CrossRef]
- Spatarelu, C.P.; Chiriac, A.L.; Cursaru, B.; Iordache, T.V.; Gavrila, A.M.; Cojocaru, C.T.; Botez, R.E.; Trica, B.; Sarbu, A.; Teodorescu, M.; et al. Composite Nanogels Based on Zeolite-Poly (ethylene glycol) Diacrylate for Controlled Drug Delivery. Nanomaterials 2020, 10, 195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raemdonck, K.; Demeester, J.; De Smedt, S. Advanced nanogel engineering for drug delivery. Soft Matter 2009, 5, 707–715. [Google Scholar] [CrossRef]
- Imani, M.; Sharifi, S.; Mirzade, H.; Ziaee, F. Monitoring of Polyethylene Glycol-diacrylate-based Hydrogel Formation by Real Time NMR Spectroscopy. Iran. Polym. J. 2007, 16, 13–20. [Google Scholar]
- Askari, F.; Zandi, M.; Shokrolahi, P.; Tabatabaei, M.H.; Hajirasoliha, E. Reduction in protein absorption on ophthalmic lenses by PEGDA bulk modifcation of silicone acrylate based formulation. Prog. Biomater. 2019, 8, 169–183. [Google Scholar] [CrossRef] [Green Version]
- Bae, M.; Gemeinhart, R.A.; Divan, R.; Suthar, K.J.; Mancini, D.C. Fabrication of Poly (ethylene glycol) Hydrogel Structures for Pharmaceutical Applications using Electron beam and Optical Lithography. J. Vac. Sci. Technol. B 2010, 28, C6P24–C6P29. [Google Scholar] [CrossRef] [Green Version]
- Goormaghtigh, E.; Cabiaux, V.; Ruysschaert, J.-M. Determination of Soluble and Membrane Protein Structure by Fourier Transform Infrared Spectroscopy. In Physicochemical Methods in the Study of Biomembranes; Subcellular Biochemistry; Hilderson, H.J., Ralston, G.B., Eds.; Springer: Boston, MA, USA, 1994; Volume 23, pp. 405–450. [Google Scholar] [CrossRef]
- Naumann, D. Ft-Infrared and Ft-Raman Spectroscopy in Biomedical Research. Appl. Spectrosc. Rev. 2001, 36, 239–298. [Google Scholar] [CrossRef]
- Kennedy, D.F.; Slotboom, A.J.; de Haas, G.H.; Chapman, D. A Fourier transform infrared spectroscopic (FTIR) study of porcine and bovine pancreatic phospholipase A2 and their interaction with substrate analogues and a transition-state inhibitor. Biochim. Biophys. Acta (BBA) Protein Struct. Molec. Enzym. 1990, 1040, 317–326. [Google Scholar] [CrossRef]
- Barth, A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta Bioenerg. 2007, 1767, 1073–1101. [Google Scholar] [CrossRef]
- Ekomo, V.M.; Branger, C.; Bikanga, R.; Florea, A.-M.; Istamboulie, G.; Calas-Blanchard, C.; Noguer, T.; Sarbu, A.; Brisset, H. Detection of Bisphenol A in aqueous medium by screen printed carbon electrodes incorporating electrochemical molecularly imprinted polymers. Biosens. Bioelectron. 2018, 112, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Barrera, F.N. On ‘Fourier transform infrared study of proteins with parallel β-chains’ by Heino Susi, D. Michael Byler. Arch. Biochem. Biophys. 2022, 726, 109114. [Google Scholar] [CrossRef] [PubMed]
- Genier, F.S.; Burdin, C.V.; Biria, S.; Hosein, I.D. A novel calcium-ion solid polymer electrolyte based on crosslinked poly (ethylene glycol) diacrylate. J. Power Sources 2019, 414, 302–307. [Google Scholar] [CrossRef]
- Maitlo, I.; Ali, S.; Akram, M.Y.; Shehzad, F.K.; Nie, J. Binary phase solid-state photopolymerization of acrylates: Design, characterization and biomineralization of 3D scaffolds for tissue engineering. Front. Mater. Sci. 2017, 11, 307–317. [Google Scholar] [CrossRef]
- Saimani, S.; Kumar, A. Polyethylene glycol diacrylate and thermoplastic polymers based semi-IPNs for asymmetric membranes. Compos. Interfaces 2008, 15, 781–797. [Google Scholar] [CrossRef] [Green Version]
- Richmond-Aylor, A.; Bell, S.; Callery, P.; Morris, K. Thermal degradation analysis of amino acids in fingerprint residue by pyrolysis GC–MS to develop new latent fingerprint developing reagents. J. Forensic Sci. 2007, 52, 380–382. [Google Scholar] [CrossRef]
- Koch, S.J.; Renner, C.; Xie, X.; Schrader, T. Tuning Linear Copolymers into Protein-Specific Hosts. Angew. Chem. Int. Ed. Engl. 2006, 118, 6500–6503. [Google Scholar] [CrossRef]
- Murakami, M.; Sato, H.; Taketomi, Y. Updating Phospholipase A2 Biology. Biomolecules 2020, 10, 1457. [Google Scholar] [CrossRef]
- Shipolini, R.A.; Callewaert, G.L.; Cottrell, R.C.; Doonan, S.; Vernon, C.A.; Banks, B.E.C. Phospholipase A From Bee Venom. Eur. J. Biochem. 1971, 20, 459–468. [Google Scholar] [CrossRef]
- Hawkins, D.M.; Ellis, E.A.; Stevenson, D.; Holzenburg, A.; Reddy, S.M. Novel critical point drying (CPD) based preparation and transmission electron microscopy (TEM) imaging of protein specific molecularly imprinted polymers (HydroMIPs). J. Mater. Sci. 2007, 42, 9465–9946. [Google Scholar] [CrossRef]
- Shi, H.; Ratner, B.D. Template recognition of protein-imprinted polymer surfaces. J. Biomed. Mater. Res. 2000, 49, 1–11. [Google Scholar] [CrossRef]
- Liu, G.; Li, Y.; Yang, L.; Wei, Y.; Wang, X.; Wang, Z.; Tao, L. Cytotoxicity study of polyethylene glycol derivatives. RSC Adv. 2017, 7, 18252–18259. [Google Scholar] [CrossRef]







| Samples | PEGDA700/ PEGDA2000 (%) | Span 80/ Tween 60 (%) | Emulsifiers/ Aqueous Phase (%) | Solvent/ Aqueous Phase (%) | PLA2/ PEGDA (Molar Ratio) |
|---|---|---|---|---|---|
| NIP-LFNG | 75/25 | 87.5:12.5 | 3 | 5.11 | 0 |
| MIP-LFNG (W) | 75/25 | 87.5:12.5 | 3 | 5.11 | 1/5 (PLA in H2O) |
| MIP-LFNG (T) | 75/25 | 87.5:12.5 | 3 | 5.11 | 1/5 (PLA in TRIS/HCl) |
| Sample | Diameter * (nm) | Polydispersity Index (PDI) |
|---|---|---|
| NIP-LFNG | 143 ± 0.53 | 0.326 |
| MIP-LFNG (W, ext) | 163 ± 2.90 | 0.251 |
| MIP-LFNG (W) | 198 ± 3.91 | 0.375 |
| MIP-LFNG (T, ext) | 170 ± 1.22 | 0.184 |
| MIP-LFNG (T) | 189 ± 3.91 | 0.322 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaharia, A.; Gavrila, A.-M.; Caras, I.; Trica, B.; Chiriac, A.-L.; Gifu, C.I.; Neblea, I.E.; Stoica, E.-B.; Dolana, S.V.; Iordache, T.-V. Molecularly Imprinted Ligand-Free Nanogels for Recognizing Bee Venom-Originated Phospholipase A2 Enzyme. Polymers 2022, 14, 4200. https://0-doi-org.brum.beds.ac.uk/10.3390/polym14194200
Zaharia A, Gavrila A-M, Caras I, Trica B, Chiriac A-L, Gifu CI, Neblea IE, Stoica E-B, Dolana SV, Iordache T-V. Molecularly Imprinted Ligand-Free Nanogels for Recognizing Bee Venom-Originated Phospholipase A2 Enzyme. Polymers. 2022; 14(19):4200. https://0-doi-org.brum.beds.ac.uk/10.3390/polym14194200
Chicago/Turabian StyleZaharia, Anamaria, Ana-Mihaela Gavrila, Iuliana Caras, Bogdan Trica, Anita-Laura Chiriac, Catalina Ioana Gifu, Iulia Elena Neblea, Elena-Bianca Stoica, Sorin Viorel Dolana, and Tanta-Verona Iordache. 2022. "Molecularly Imprinted Ligand-Free Nanogels for Recognizing Bee Venom-Originated Phospholipase A2 Enzyme" Polymers 14, no. 19: 4200. https://0-doi-org.brum.beds.ac.uk/10.3390/polym14194200