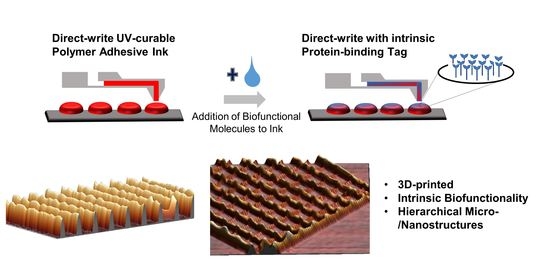
Integration of Biofunctional Molecules into 3D-Printed Polymeric Micro-/Nanostructures
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. FluidFM Printing
2.3. Characterization of Printed Structures with Atomic Force Microscopy (AFM)
2.4. Protein Binding
2.5. Optical Microscopy
2.6. Substrate Functionalization
2.7. Characterization of Substrate Wettability
2.8. Statistical Analysis
3. Results
3.1. Printing of Pure Adhesive
3.2. Printing with Functionalized Adhesive
3.2.1. Preparation of Functionalized Adhesive Inks
3.2.2. Patterning on Substrates with Different Wettability
3.2.3. Comparison of Mechanical Properties
3.3. Biofunctionalization/Protein Binding
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chung, J.J.; Im, H.; Kim, S.H.; Park, J.W.; Jung, Y. Toward Biomimetic Scaffolds for Tissue Engineering: 3D Printing Techniques in Regenerative Medicine. Front. Bioeng. Biotechnol. 2020, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Hippler, M.; Lemma, E.D.; Bertels, S.; Blasco, E.; Barner-Kowollik, C.; Wegener, M.; Bastmeyer, M. 3D Scaffolds to Study Basic Cell Biology. Adv. Mater. 2019, 31, 1808110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ouyang, X.; Zhang, K.; Wu, J.; Wong, D.S.-H.; Feng, Q.; Bian, L.; Zhang, A.P. Optical µ-Printing of Cellular-Scale Microscaffold Arrays for 3D Cell Culture. Sci. Rep. 2017, 7, 8880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zandrini, T.; Shan, O.; Parodi, V.; Cerullo, G.; Raimondi, M.T.; Osellame, R. Multi-foci laser microfabrication of 3D polymeric scaffolds for stem cell expansion in regenerative medicine. Sci. Rep. 2019, 9, 11761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richter, B.; Hahn, V.; Bertels, S.; Claus, T.K.; Wegener, M.; Delaittre, G.; Barner-Kowollik, C.; Bastmeyer, M. Guiding Cell Attachment in 3D Microscaffolds Selectively Functionalized with Two Distinct Adhesion Proteins. Adv. Mater. 2017, 29, 1604342. [Google Scholar] [CrossRef] [PubMed]
- Barner-Kowollik, C.; Bastmeyer, M.; Blasco, E.; Delaittre, G.; Müller, P.; Richter, B.; Wegener, M. 3D Laser Micro- and Nanoprinting: Challenges for Chemistry. Angew. Chem. Int. Ed. 2017, 56, 15828–15845. [Google Scholar] [CrossRef] [PubMed]
- Bettinger, C.J.; Langer, R.; Borenstein, J.T. Engineering Substrate Topography at the Micro- and Nanoscale to Control Cell Function. Angew. Chem. Int. Ed. 2009, 48, 5406–5415. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.; Du, Z.; Qiu, H.; Gao, P.; Zhao, X.; Wang, H.; Tu, Q.; Xiong, K.; Huang, N.; Yang, Z. From surface to bulk modification: Plasma polymerization of amine-bearing coating by synergic strategy of biomolecule grafting and nitric oxide loading. Bioact. Mater. 2020, 5, 17–25. [Google Scholar] [CrossRef]
- Ferrari, M.; Cirisano, F.; Morán, M.C. Mammalian Cell Behavior on Hydrophobic Substrates: Influence of Surface Properties. Colloids Interfaces 2019, 3, 48. [Google Scholar] [CrossRef] [Green Version]
- Apte, G.; Lindenbauer, A.; Schemberg, J.; Rothe, H.; Nguyen, T.H. Controlling Surface-Induced Platelet Activation by Agarose and Gelatin-Based Hydrogel Films. ACS Omega 2021, 6, 10963–10974. [Google Scholar] [CrossRef]
- Bui, V.-C.; Medvedev, N.; Apte, G.; Chen, L.-Y.; Denker, C.; Greinacher, A.; Nguyen, T.-H. Response of Human Blood Platelets on Nanoscale Groove Patterns: Implications for Platelet Storage. ACS Appl. Nano Mater. 2020, 3, 6996–7004. [Google Scholar] [CrossRef]
- Apte, G.; Börke, J.; Rothe, H.; Liefeith, K.; Nguyen, T.H. Modulation of Platelet-Surface Activation: Current State and Future Perspectives. ACS Appl. Bio Mater. 2020, 3, 5574–5589. [Google Scholar] [CrossRef] [PubMed]
- Hauptmann, N.; Lian, Q.; Ludolph, J.; Rothe, H.; Hildebrand, G.; Liefeith, K. Biomimetic Designer Scaffolds Made of D,L-Lactide-ε-Caprolactone Polymers by 2-Photon Polymerization. Tissue Eng. Part B Rev. 2019, 25, 167–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulangara, K.; Leong, K.W. Substrate topography shapes cell function. Soft Matter 2009, 5, 4072. [Google Scholar] [CrossRef]
- Xiang, T.; Hou, J.; Xie, H.; Liu, X.; Gong, T.; Zhou, S. Biomimetic micro/nano structures for biomedical applications. Nano Today 2020, 35, 100980. [Google Scholar] [CrossRef]
- Zambelli, T.; Aebersold, M.; Behr, P.; Han, H.; Hirt, L.; Guillaume-gentil, O.; Vörös, J. FluidFM: Development of the Instrument as well as Its Applications for 2D and 3D Lithography. In Open-Space Microfluidics: Concepts, Implementation, and Applications; Wiley-VCH Verlag: Weinheim, Germany, 2018; pp. 295–322. [Google Scholar] [CrossRef]
- Grüter, R.R.; Vörös, J.; Zambelli, T. FluidFM as a lithography tool in liquid: Spatially controlled deposition of fluorescent nanoparticles. Nanoscale 2013, 5, 1097–1104. [Google Scholar] [CrossRef]
- Berganza, E.; Hirtz, M. Direct-Write Patterning of Biomimetic Lipid Membranes In Situ with FluidFM. ACS Appl. Mater. Interfaces 2021, 13, 50774–50784. [Google Scholar] [CrossRef]
- Ventrici de Souza, J.; Liu, Y.; Wang, S.; Dörig, P.; Kuhl, T.L.; Frommer, J.; Liu, G. Three-Dimensional Nanoprinting via Direct Delivery. J. Phys. Chem. B 2018, 122, 956–962. [Google Scholar] [CrossRef] [Green Version]
- Deng, W.N.; Wang, S.; Ventrici de Souza, J.; Kuhl, T.L.; Liu, G. New Algorithm to Enable Construction and Display of 3D Structures from Scanning Probe Microscopy Images Acquired Layer-by-Layer. J. Phys. Chem. A 2018, 122, 5756–5763. [Google Scholar] [CrossRef]
- Henkel Technical Data Sheet—Loctite® AA 3491TM. 2014. Available online: https://www.henkel-adhesives.com/de/de/produkt/uv-curing-adhesives/loctite_aa_3491.html (accessed on 24 February 2022).
- Meyer, R.; Giselbrecht, S.; Rapp, B.E.; Hirtz, M.; Niemeyer, C.M. Advances in DNA-directed immobilization. Curr. Opin. Chem. Biol. 2014, 18, 8–15. [Google Scholar] [CrossRef]
- Kumar, R.; Weigel, S.; Meyer, R.; Niemeyer, C.M.; Fuchs, H.; Hirtz, M. Multi-color polymer pen lithography for oligonucleotide arrays. Chem. Commun. 2016, 52, 12310–12313. [Google Scholar] [CrossRef] [PubMed]
- Horcas, I.; Fernández, R.; Gómez-Rodríguez, J.M.; Colchero, J.; Gómez-Herrero, J.; Baro, A.M. WSXM: A software for scanning probe microscopy and a tool for nanotechnology. Rev. Sci. Instrum. 2007, 78, 013705. [Google Scholar] [CrossRef] [PubMed]
- Wilchek, M.; Bayer, E.A. Introduction to avidin-biotin technology. In Methods in Enzymology—Vol. 184: Avidin-Biotin Technology; Academic Press: Cambridge, MA, USA, 1990; pp. 5–13. [Google Scholar] [CrossRef]
- Liu, H.-Y.; Kumar, R.; Zhong, C.; Gorji, S.; Paniushkina, L.; Masood, R.; Wittel, U.A.; Fuchs, H.; Nazarenko, I.; Hirtz, M. Rapid Capture of Cancer Extracellular Vesicles by Lipid Patch Microarrays. Adv. Mater. 2021, 33, 2008493. [Google Scholar] [CrossRef] [PubMed]
- Padmanabhan, J.; Kinser, E.R.; Stalter, M.A.; Duncan-Lewis, C.; Balestrini, J.L.; Sawyer, A.J.; Schroers, J.; Kyriakides, T.R. Engineering Cellular Response Using Nanopatterned Bulk Metallic Glass. ACS Nano 2014, 8, 4366–4375. [Google Scholar] [CrossRef] [PubMed]
- Cyster, L.A.; Parker, K.G.; Parker, T.L.; Grant, D.M. The effect of surface chemistry and nanotopography of titanium nitride (TiN) films on primary hippocampal neurones. Biomaterials 2004, 25, 97–107. [Google Scholar] [CrossRef]
- Salloum, D.S.; Olenych, S.G.; Keller, T.C.S.; Schlenoff, J.B. Vascular Smooth Muscle Cells on Polyelectrolyte Multilayers: Hydrophobicity-Directed Adhesion and Growth. Biomacromolecules 2005, 6, 161–167. [Google Scholar] [CrossRef]
- Tan, G.; Ouyang, K.; Wang, H.; Zhou, L.; Wang, X.; Liu, Y.; Zhang, L.; Ning, C. Effect of Amino-, Methyl- and Epoxy-Silane Coupling as a Molecular Bridge for Formatting a Biomimetic Hydroxyapatite Coating on Titanium by Electrochemical Deposition. J. Mater. Sci. Technol. 2016, 32, 956–965. [Google Scholar] [CrossRef]
- Breuls, R.G.M.; Jiya, T.U.; Smit, T.H. Scaffold Stiffness Influences Cell Behavior: Opportunities for Skeletal Tissue Engineering. Open Orthop. J. 2008, 2, 103–109. [Google Scholar] [CrossRef] [Green Version]
- Gautam, R.; Singh, R.D.; Sharma, V.P.; Siddhartha, R.; Chand, P.; Kumar, R. Biocompatibility of polymethylmethacrylate resins used in dentistry. J. Biomed. Mater. Res. Part B Appl. Biomater. 2012, 100B, 1444–1450. [Google Scholar] [CrossRef]
- Alifui-Segbaya, F.; George, R. Biocompatibility of 3d-printed methacrylate for hearing devices. Inventions 2018, 3, 52. [Google Scholar] [CrossRef] [Green Version]





| Substrate Label | S1 | S2 | S3 | S4 |
|---|---|---|---|---|
| Functionalization | 7-octenyl trichlorosilane | None | (3-glycidyl oxypropyl)-trimethoxysilan | O2 plasma activation |
| Width * (nm) | - ** | 300 ± 21 | 524 ± 26 | 1457 ± 65 |
| Height (nm) | - ** | 68 ± 16 | 32 ± 3 | 15 ± 1 |
| Aspect ratio *** | - | 0.23 ± 0.04 | 0.06 ± 0.01 | 0.01 ± 0.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berganza, E.; Apte, G.; Vasantham, S.K.; Nguyen, T.-H.; Hirtz, M. Integration of Biofunctional Molecules into 3D-Printed Polymeric Micro-/Nanostructures. Polymers 2022, 14, 1327. https://0-doi-org.brum.beds.ac.uk/10.3390/polym14071327
Berganza E, Apte G, Vasantham SK, Nguyen T-H, Hirtz M. Integration of Biofunctional Molecules into 3D-Printed Polymeric Micro-/Nanostructures. Polymers. 2022; 14(7):1327. https://0-doi-org.brum.beds.ac.uk/10.3390/polym14071327
Chicago/Turabian StyleBerganza, Eider, Gurunath Apte, Srivatsan K. Vasantham, Thi-Huong Nguyen, and Michael Hirtz. 2022. "Integration of Biofunctional Molecules into 3D-Printed Polymeric Micro-/Nanostructures" Polymers 14, no. 7: 1327. https://0-doi-org.brum.beds.ac.uk/10.3390/polym14071327