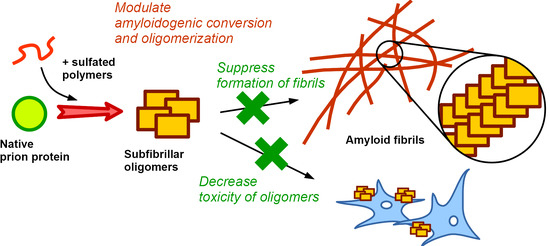
Synthetic Sulfated Polymers Control Amyloid Aggregation of Ovine Prion Protein and Decrease Its Toxicity
Abstract
:1. Introduction
2. Materials and Methods
2.1. PrP Purification
2.2. Polymers
2.3. PrP Aggregation Assay
2.4. DLS
2.5. Thioflavin T Fluorescence Measurements
2.6. Cytotoxicity Assay
2.7. Statistical Analysis
2.8. MD Simulations
3. Results
3.1. Amyloid Conversion and Oligomerization of Prion Protein
3.2. Modeling of Protein–Polymer Interaction
3.3. Formation of Fibrils
3.4. Amyloidogenic Capacity of Fibrils (Seeding)
3.5. Cytotoxicity of PrP Oligomers
4. Discussion and Conclusion Remarks
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sipe, J.D.; Benson, M.D.; Buxbaum, J.N.; Ikeda, S.; Merlini, G.; Saraiva, M.J.M.; Westermark, P. Amyloid Fibril Proteins and Amyloidosis: Chemical Identification and Clinical Classification International Society of Amyloidosis 2016 Nomenclature Guidelines. Amyloid 2016, 23, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Iadanza, M.G.; Jackson, M.P.; Hewitt, E.W.; Ranson, N.A.; Radford, S.E. A New Era for Understanding Amyloid Structures and Disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 755–773. [Google Scholar] [CrossRef] [PubMed]
- Aguzzi, A.; Calella, A.M. Prions: Protein Aggregation and Infectious Diseases. Physiol. Rev. 2009, 89, 1105–1152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguzzi, A.; Sigurdson, C.; Heikenwaelder, M. Molecular Mechanisms of Prion Pathogenesis. Annu. Rev. Pathol. Mech. Dis. 2008, 3, 11–40. [Google Scholar] [CrossRef]
- Prusiner, S.B. Neurodegenerative Diseases and Prions. N. Engl. J. Med. 2001, 344, 1516–1526. [Google Scholar] [CrossRef]
- Lazarev, V.F.; Mikhaylova, E.R.; Guzhova, I.V.; Margulis, B.A. Possible Function of Molecular Chaperones in Diseases Caused by Propagating Amyloid Aggregates. Front. Neurosci. 2017, 11, 277. [Google Scholar] [CrossRef] [Green Version]
- Paul, S.; Mahanta, S. Association of Heat-Shock Proteins in Various Neurodegenerative Disorders: Is It a Master Key to Open the Therapeutic Door? Mol. Cell. Biochem. 2014, 386, 45–61. [Google Scholar] [CrossRef]
- Chernoff, Y.O.; Lindquist, S.L.; Ono, B.; Inge-Vechtomov, S.G.; Liebman, S.W. Role of the Chaperone Protein Hsp104 in Propagation of the Yeast Prion-like Factor [Psi+]. Science 1995, 268, 880–884. [Google Scholar] [CrossRef]
- Krobitsch, S.; Lindquist, S. Aggregation of Huntingtin in Yeast Varies with the Length of the Polyglutamine Expansion and the Expression of Chaperone Proteins. Proc. Natl. Acad. Sci. USA 2000, 97, 1589–1594. [Google Scholar] [CrossRef] [Green Version]
- Newnam, G.P.; Wegrzyn, R.D.; Lindquist, S.L.; Chernoff, Y.O. Antagonistic Interactions between Yeast Chaperones Hsp104 and Hsp70 in Prion Curing. Mol. Cell. Biol. 1999, 19, 1325–1333. [Google Scholar] [CrossRef] [Green Version]
- Bobkova, N.V.; Garbuz, D.G.; Nesterova, I.; Medvinskaya, N.; Samokhin, A.; Alexandrova, I.; Yashin, V.; Karpov, V.; Kukharsky, M.S.; Ninkina, N.N.; et al. Therapeutic Effect of Exogenous Hsp70 in Mouse Models of Alzheimer’s Disease. J. Alzheimer’s Dis. 2014, 38, 425–435. [Google Scholar] [CrossRef] [Green Version]
- Semenyuk, P.; Tiainen, T.; Hietala, S.; Tenhu, H.; Aseyev, V.; Muronetz, V. Artificial Chaperones Based on Thermoresponsive Polymers Recognize the Unfolded State of the Protein. Int. J. Biol. Macromol. 2019, 121, 536–545. [Google Scholar] [CrossRef] [Green Version]
- Nomura, Y.; Sasaki, Y.; Takagi, M.; Narita, T.; Aoyama, Y.; Akiyoshi, K. Thermoresponsive Controlled Association of Protein with a Dynamic Nanogel of Hydrophobized Polysaccharide and Cyclodextrin: Heat Shock Protein-Like Activity of Artificial Molecular Chaperone. Biomacromolecules 2005, 6, 447–452. [Google Scholar] [CrossRef]
- Martin, N.; Ma, D.; Herbet, A.; Boquet, D.; Winnik, F.M.; Tribet, C. Prevention of Thermally Induced Aggregation of IgG Antibodies by Noncovalent Interaction with Poly(Acrylate) Derivatives. Biomacromolecules 2014, 15, 2952–2962. [Google Scholar] [CrossRef]
- Martin, N.; Ruchmann, J.; Tribet, C. Prevention of Aggregation and Renaturation of Carbonic Anhydrase via Weak Association with Octadecyl- or Azobenzene-Modified Poly(Acrylate) Derivatives. Langmuir 2015, 31, 338–349. [Google Scholar] [CrossRef]
- Shalova, I.N.; Asryants, R.A.; Sholukh, M.V.; Saso, L.; Kurganov, B.I.; Muronetz, V.I.; Izumrudov, V.A. Interaction of Polyanions with Basic Proteins, 2(a): Influence of Complexing Polyanions on the Thermo-Aggregation of Oligomeric Enzymes. Macromolar Biosci. 2005, 5, 1184–1192. [Google Scholar] [CrossRef]
- Semenyuk, P.I.; Moiseeva, E.V.; Stroylova, Y.Y.; Lotti, M.; Izumrudov, V.A.; Muronetz, V.I. Sulfated and Sulfonated Polymers Are Able to Solubilize Efficiently the Protein Aggregates of Different Nature. Arch. Biochem. Biophys. 2015, 567, 22–29. [Google Scholar] [CrossRef]
- Klajnert, B.; Cladera, J.; Bryszewska, M. Molecular Interactions of Dendrimers with Amyloid Peptides: PH Dependence. Biomacromolecules 2006, 7, 2186–2191. [Google Scholar] [CrossRef]
- Klajnert, B.; Cortijo-Arellano, M.; Cladera, J.; Bryszewska, M. Influence of Dendrimer’s Structure on Its Activity against Amyloid Fibril Formation. Biochem. Biophys. Res. Commun. 2006, 345, 21–28. [Google Scholar] [CrossRef]
- Van Horssen, J.; Wesseling, P.; van den Heuvel, L.P.; de Waal, R.M.; Verbeek, M.M. Heparan Sulphate Proteoglycans in Alzheimer’s Disease and Amyloid-related Disorders. Lancet Neurol. 2003, 2, 482–492. [Google Scholar] [CrossRef]
- Iannuzzi, C.; Irace, G.; Sirangelo, I. The Effect of Glycosaminoglycans (GAGs) on Amyloid Aggregation and Toxicity. Molecules 2015, 20, 2510–2528. [Google Scholar] [CrossRef] [Green Version]
- Nishitsuji, K.; Uchimura, K. Sulfated Glycosaminoglycans in Protein Aggregation Diseases. Glycoconj. J. 2017, 34, 453–466. [Google Scholar] [CrossRef]
- McLaurin, J.; Franklin, T.; Zhang, X.; Deng, J.; Fraser, P.E. Interactions of Alzheimer Amyloid-β Peptides with Glycosaminoglycans. Eur. J. Biochem. 1999, 266, 1101–1110. [Google Scholar] [CrossRef]
- Timmer, N.M.; Schirris, T.J.J.; Bruinsma, I.B.; Otte-Höller, I.; van Kuppevelt, T.H.; de Waal, R.M.W.; Verbeek, M.M. Aggregation and Cytotoxic Properties towards Cultured Cerebrovascular Cells of Dutch-Mutated Aβ40 (DAβ1-40) Are Modulated by Sulfate Moieties of Heparin. Neurosci. Res. 2010, 66, 380–389. [Google Scholar] [CrossRef]
- Cohlberg, J.A.; Li, J.; Uversky, V.N.; Fink, A.L. Heparin and Other Glycosaminoglycans Stimulate the Formation of Amyloid Fibrils from α-Synuclein in Vitro. Biochemistry 2002, 41, 1502–1511. [Google Scholar] [CrossRef]
- Hasegawa, M.; Crowther, R.A.; Jakes, R.; Goedert, M. Alzheimer-like Changes in Microtubule-Associated Protein Tau Induced by Sulfated Glycosaminoglycans. Inhibition of Microtubule Binding, Stimulation of Phosphorylation, and Filament Assembly Depend on the Degree of Sulfation. J. Biol. Chem. 1997, 272, 33118–33124. [Google Scholar] [CrossRef] [Green Version]
- Rosú, S.A.; Toledo, L.; Urbano, B.F.; Sanchez, S.A.; Calabrese, G.C.; Tricerri, M.A. Learning from Synthetic Models of Extracellular Matrix; Differential Binding of Wild Type and Amyloidogenic Human Apolipoprotein A-I to Hydrogels Formed from Molecules Having Charges Similar to Those Found in Natural GAGs. Protein J. 2017, 36, 374–383. [Google Scholar] [CrossRef]
- Townsend, D.; Hughes, E.; Hussain, R.; Siligardi, G.; Baldock, S.; Madine, J.; Middleton, D.A. Heparin and Methionine Oxidation Promote the Formation of Apolipoprotein A-I Amyloid Comprising α-Helical and β-Sheet Structures. Biochemistry 2017, 56, 1632–1644. [Google Scholar] [CrossRef] [PubMed]
- Solomon, J.P.; Bourgault, S.; Powers, E.T.; Kelly, J.W. Heparin Binds 8 KDa Gelsolin Cross-β-Sheet Oligomers and Accelerates Amyloidogenesis by Hastening Fibril Extension. Biochemistry 2011, 50, 2486–2498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, M.; Kawakami, T.; Okino, N.; Sasaki, K.; Nakanishi, K.; Takase, H.; Yamada, T.; Mukai, T. Acceleration of Amyloid Fibril Formation by Carboxyl-Terminal Truncation of Human Serum Amyloid A. Arch. Biochem. Biophys. 2018, 639, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Semenyuk, P.; Kurochkina, L.; Barinova, K.; Muronetz, V. Alpha-Synuclein Amyloid Aggregation Is Inhibited by Sulfated Aromatic Polymers and Pyridinium Polycation. Polymers 2020, 12, 517. [Google Scholar] [CrossRef] [Green Version]
- Caughey, B.; Raymond, G.J. Sulfated Polyanion Inhibition of Scrapie-Associated PrP Accumulation in Cultured Cells. J. Virol. 1993, 67, 643–650. [Google Scholar] [CrossRef] [Green Version]
- Evstafyeva, D.B.; Izumrudov, V.A.; Muronetz, V.I.; Semenyuk, P.I. Tightly Bound Polyelectrolytes Enhance Enzyme Proteolysis and Destroy Amyloid Aggregates. Soft Matter 2018, 14, 3768–3773. [Google Scholar] [CrossRef]
- Semenyuk, P.; Muronetz, V. Protein Interaction with Charged Macromolecules: From Model Polymers to Unfolded Proteins and Post-Translational Modifications. Int. J. Mol. Sci. 2019, 20, 1252. [Google Scholar] [CrossRef] [Green Version]
- Achazi, K.; Haag, R.; Ballauff, M.; Dernedde, J.; Kizhakkedathu, J.N.; Maysinger, D.; Multhaup, G. Understanding the Interaction of Polyelectrolyte Architectures with Proteins and Biosystems. Angew. Chem. Int. Ed. 2021, 60, 3882–3904. [Google Scholar] [CrossRef]
- Chakroun, N.; Fornili, A.; Prigent, S.; Kleinjung, J.; Dreiss, C.A.; Rezaei, H.; Fraternali, F. Decrypting Prion Protein Conversion into a β-Rich Conformer by Molecular Dynamics. J. Chem. Theory Comput. 2013, 9, 2455–2465. [Google Scholar] [CrossRef]
- Huy, P.D.Q.; Vuong, Q.V.; La Penna, G.; Faller, P.; Li, M.S. Impact of Cu(II) Binding on Structures and Dynamics of Aβ42 Monomer and Dimer: Molecular Dynamics Study. ACS Chem. Neurosci. 2016, 7, 1348–1363. [Google Scholar] [CrossRef]
- Mezentsev, Y.V.; Medvedev, A.E.; Kechko, O.I.; Makarov, A.A.; Ivanov, A.S.; Mantsyzov, A.B.; Kozin, S.A. Zinc-Induced Heterodimer Formation between Metal-Binding Domains of Intact and Naturally Modified Amyloid-Beta Species: Implication to Amyloid Seeding in Alzheimer’s Disease? J. Biomol. Struct. Dyn. 2016, 34, 2317–2326. [Google Scholar] [CrossRef]
- Nasica-Labouze, J.; Nguyen, P.H.; Sterpone, F.; Berthoumieu, O.; Buchete, N.-V.; Coté, S.; De Simone, A.; Doig, A.J.; Faller, P.; Garcia, A.; et al. Amyloid β Protein and Alzheimer’s Disease: When Computer Simulations Complement Experimental Studies. Chem. Rev. 2015, 115, 3518–3563. [Google Scholar] [CrossRef]
- Verma, M.; Vats, A.; Taneja, V. Toxic Species in Amyloid Disorders: Oligomers or Mature Fibrils. Ann. Indian Acad. Neurol. 2015, 18, 138–145. [Google Scholar] [CrossRef]
- Bengoa-Vergniory, N.; Roberts, R.F.; Wade-Martins, R.; Alegre-Abarrategui, J. Alpha-Synuclein Oligomers: A New Hope. Acta Neuropathol. 2017, 134, 819–838. [Google Scholar] [CrossRef] [Green Version]
- Soto, C.; Estrada, L.D. Protein Misfolding and Neurodegeneration. Arch. Neurol. 2008, 65, 184–189. [Google Scholar] [CrossRef] [Green Version]
- Rezaei, H.; Marc, D.; Choiset, Y.; Takahashi, M.; Hui Bon Hoa, G.; Haertlé, T.; Grosclaude, J.; Debey, P. High Yield Purification and Physico-Chemical Properties of Full-Length Recombinant Allelic Variants of Sheep Prion Protein Linked to Scrapie Susceptibility. Eur. J. Biochem. 2000, 267, 2833–2839. [Google Scholar] [CrossRef]
- Rezaei, H.; Eghiaian, F.; Perez, J.; Doublet, B.; Choiset, Y.; Haertle, T.; Grosclaude, J. Sequential Generation of Two Structurally Distinct Ovine Prion Protein Soluble Oligomers Displaying Different Biochemical Reactivities. J. Mol. Biol. 2005, 347, 665–679. [Google Scholar] [CrossRef]
- Wilham, J.M.; Orrú, C.D.; Bessen, R.A.; Atarashi, R.; Sano, K.; Race, B.; Meade-White, K.D.; Taubner, L.M.; Timmes, A.; Caughey, B. Rapid End-Point Quantitation of Prion Seeding Activity with Sensitivity Comparable to Bioassays. PLoS Pathog. 2010, 6, e1001217. [Google Scholar] [CrossRef] [Green Version]
- Dupradeau, F.-Y.; Pigache, A.; Zaffran, T.; Savineau, C.; Lelong, R.; Grivel, N.; Lelong, D.; Rosanski, W.; Cieplak, P. The R.E.D. Tools: Advances in RESP and ESP Charge Derivation and Force Field Library Building. Phys. Chem. Chem. Phys. 2010, 12, 7821. [Google Scholar] [CrossRef] [Green Version]
- Granovsky, A.A. Firefly Version 8.0. Available online: http://classic.chem.msu.su/gran/gamess/index.html (accessed on 26 May 2016).
- Schmidt, M.W.; Baldridge, K.K.; Boatz, J.A.; Elbert, S.T.; Gordon, M.S.; Jensen, J.H.; Koseki, S.; Matsunaga, N.; Nguyen, K.A.; Su, S.; et al. General Atomic and Molecular Electronic Structure System. J. Comput. Chem. 1993, 14, 1347–1363. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular Dynamics with Coupling to an External Bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef] [Green Version]
- Stogov, S.V.; Izumrudov, V.A.; Muronetz, V.I. Structural Changes of a Protein Bound to a Polyelectrolyte Depend on the Hydrophobicity and Polymerization Degree of the Polyelectrolyte. Biochem. Mosc. 2010, 75, 437–442. [Google Scholar] [CrossRef]
- Yeh, V.; Broering, J.M.; Romanyuk, A.; Chen, B.; Chernoff, Y.O.; Bommarius, A.S. The Hofmeister Effect on Amyloid Formation Using Yeast Prion Protein. Protein Sci. Publ. Protein Soc. 2010, 19, 47–56. [Google Scholar] [CrossRef] [Green Version]
- Castillo, G.M.; Lukito, W.; Wight, T.N.; Snow, A.D. The Sulfate Moieties of Glycosaminoglycans Are Critical for the Enhancement of β-Amyloid Protein Fibril Formation. J. Neurochem. 1999, 72, 1681–1687. [Google Scholar] [CrossRef] [PubMed]
- Kisilevsky, R.; Ancsin, J.B.; Szarek, W.A.; Petanceska, S. Heparan Sulfate as a Therapeutic Target in Amyloidogenesis: Prospects and Possible Complications. Amyloid 2007, 14, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Chebotareva, N.; Kurganov, B.; Livanova, N. Biochemical Effects of Molecular Crowding. Biochem. Mosc. 2004, 69, 1239. [Google Scholar] [CrossRef]
- Schreck, J.S.; Bridstrup, J.; Yuan, J.-M. Investigating the Effects of Molecular Crowding on the Kinetics of Protein Aggregation. J. Phys. Chem. B 2020, 124, 9829–9839. [Google Scholar] [CrossRef]
- Voevodin, V.V.; Zhumatiy, S.A.; Sobolev, S.I.; Antonov, A.S.; Bryzgalov, P.A.; Nikitenko, D.A.; Stefanov, K.S.; Voevodin, V.V. Practice of “Lomonosov” supercomputer. Open Syst. J. 2012, 7, 36–39. [Google Scholar]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Semenyuk, P.; Evstafyeva, D.; Izumrudov, V.; Muronetz, V. Synthetic Sulfated Polymers Control Amyloid Aggregation of Ovine Prion Protein and Decrease Its Toxicity. Polymers 2022, 14, 1478. https://0-doi-org.brum.beds.ac.uk/10.3390/polym14071478
Semenyuk P, Evstafyeva D, Izumrudov V, Muronetz V. Synthetic Sulfated Polymers Control Amyloid Aggregation of Ovine Prion Protein and Decrease Its Toxicity. Polymers. 2022; 14(7):1478. https://0-doi-org.brum.beds.ac.uk/10.3390/polym14071478
Chicago/Turabian StyleSemenyuk, Pavel, Diana Evstafyeva, Vladimir Izumrudov, and Vladimir Muronetz. 2022. "Synthetic Sulfated Polymers Control Amyloid Aggregation of Ovine Prion Protein and Decrease Its Toxicity" Polymers 14, no. 7: 1478. https://0-doi-org.brum.beds.ac.uk/10.3390/polym14071478