Analysis of Proteomic Profile of Contrasting Phosphorus Responsive Rice Cultivars Grown under Phosphorus Deficiency
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Protein Extraction
2.3. Two-Dimensional Gel Electrophoresis
2.4. Gel Analysis
2.5. In-Gel Digestion and MALDI-TOF/TOF-MS Analysis
2.6. Protein Identification and Database Searching
2.7. Statistical Analysis
3. Results
3.1. Morphological Traits as Influenced by Low-P and Optimum-P Treatments
3.2. Number of Differentially Expressed Proteins
3.3. Spatial and Functional Categorization of Differentially Expressed Proteins
3.4. Differentially Expressed Proteins of Rice Cultivars under Low P Condition
3.4.1. Common Differentially Expressed Proteins of Cv. Nagina and Cv. Panvel under Low P Condition
3.4.2. Differentially Expressed Proteins of Cv. Panvel only under low P Condition
3.4.3. Differentially Expressed Proteins of Cv. Nagina only under Low P Condition
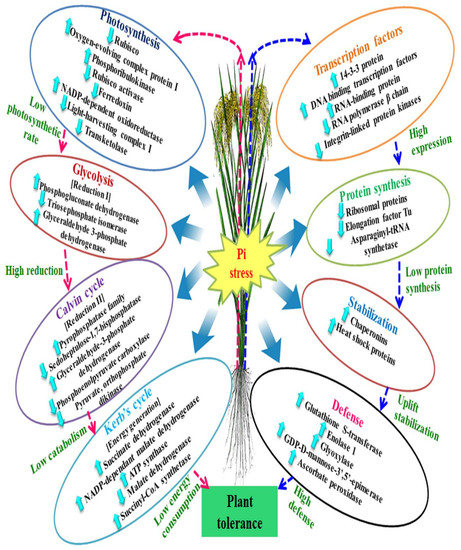
4. Discussion
4.1. Expression Pattern of the Proteins of Energy Metabolism under P-deprivation
4.2. Expression Pattern of the Proteins Involved in Transcription and Translation
4.3. Photosynthesis and CO2 Regulation under P-deficiency
4.4. Expression Proteins of Antioxidant Defense System under P-deficiency
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| PUE | Phosphate utilization efficiency |
| 2DE | Two-dimensional gel electrophoresis |
| DEP | Differentially expressed proteins |
| MALDI-TOF | Matrix-assisted laser desorption/ionization-time of flight |
References
- Fischer, R.A.; Edmeades, G.O. Breeding and Cereal Yield Progress. Crop. Sci. 2010, 50, S-85–S-98. [Google Scholar] [CrossRef] [Green Version]
- Lynch, J.P. Root Phenes for Enhanced Soil Exploration and Phosphorus Acquisition: Tools for Future Crops. Plant Physiol. 2011, 156, 1041–1049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doberman, A.; Fairhurst, T. Rice: Nutrient Disorders and Nutrient Management; IRRI & PPI & PPIC: Makati, Philippines; Singapore, 2000. [Google Scholar]
- Heuer, S.; Gaxiola, R.; Schilling, R.; Herrera-Estrella, L.; López-Arredondo, D.; Wissuwa, M.; Delhaize, E.; Rouached, H. Improving phosphorus use efficiency: A complex trait with emerging opportunities. Plant J. 2017, 90, 868–885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Kauwenbergh, S.J. World Phosphate Rock Reserves and Resources; IFDC: Muscle Shoals, AL, USA, 2010. [Google Scholar]
- Dawe, D. The potential role of biological nitrogen fixation in meeting future demand for rice and fertilizer. In The Quest for Nitrogen Fixation in Rice; Ladha, J.K., Reddy, P.M., Eds.; International Rice Research Institute: Los Banos, Philippines, 2000; pp. 93–118. [Google Scholar]
- Van Der Velde, M.; Folberth, C.; Balkovič, J.; Ciais, P.; Fritz, S.; Janssens, I.A.; Obersteiner, M.; See, L.; Skalský, R.; Xiong, W.; et al. African crop yield reductions due to increasingly unbalanced Nitrogen and Phosphorus consumption. Glob. Chang. Boil. 2014, 20, 1278–1288. [Google Scholar] [CrossRef]
- Chiera, J.; Thomas, J.; Rufty, T.W. Leaf initiation and development in soybean under phosphorus stress. J. Exp. Bot. 2002, 53, 473–481. [Google Scholar] [CrossRef] [Green Version]
- Brooks, A. Effects of Phosphorus Nutrition on Ribulose-1, 5-Bisphosphate Carboxylase Activation, Photosynthetic Quantum Yield and Amounts of Some Calvin-Cycle Metabolites in Spinach Leaves. Funct. Plant Boil. 1986, 13, 221. [Google Scholar] [CrossRef]
- Tang, X.R.; Yu, T.Q. Effects and mechanisms of P and K nutrients on yield and protein content of fodder rice. Agric. Sci. China 2002, 1, 432–437. [Google Scholar]
- Qiu, H.; Liu, C.; Yu, T.; Mei, X.; Wang, G.; Wang, J.; Cai, Y. Identification of QTL for acid phosphatase activity in root and rhizosphere soil of maize under low phosphorus stress. Euphytica 2014, 197, 133–143. [Google Scholar] [CrossRef]
- Jiang, H.; Zhang, J.; Han, Z.; Yang, J.; Ge, C.; Wu, Q. Revealing new insights into different phosphorus-starving responses between two maize (Zea mays) inbred lines by transcriptomic and proteomic studies. Sci. Rep. 2017, 7, 44294. [Google Scholar] [CrossRef] [Green Version]
- Ganie, A.H.; Ahmad, A.; Pandey, R.; Aref, I.M.; Yousuf, P.Y.; Ahmad, S.; Iqbal, M. Metabolite Profiling of Low-P Tolerant and Low-P Sensitive Maize Genotypes under Phosphorus Starvation and Restoration Conditions. PLoS ONE 2015, 10, e0129520. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Liang, W.; Sturrock, C.J.; Pandey, B.K.; Giri, J.; Mairhofer, S.; Wang, D.; Müller, L.; Tan, H.; York, L.M.; et al. Rice actin binding protein RMD controls crown root angle in response to external phosphate. Nat. Commun. 2018, 9, 2346. [Google Scholar] [CrossRef] [Green Version]
- Alexova, R.; Millar, A.H. Proteomics of phosphate use and deprivation in plants. Proteomics 2013, 13, 609–623. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.G.; Wang, Y.; Lee, C.H.; Mun, B.G.; Kim, P.J.; Lee, S.Y.; Kim, Y.C.; Kang, K.Y.; Rakwal, R.; Agrawal, G.K.; et al. A comparative proteomics survey of proteins responsive to phosphorous starvation in roots of hydroponically-grown rice seedlings. J. Korean Soc. Appl. Biol. 2011, 54, 667–677. [Google Scholar] [CrossRef]
- Torabi, S.; Wissuwa, M.; Heidari, M.; Naghavi, M.R.; Gilany, K.; Hajirezaei, M.-R.; Omidi, M.; Yazdi-Samadi, B.; Ismail, A.M.; Salekdeh, G.H. A comparative proteome approach to decipher the mechanism of rice adaptation to phosphorous deficiency. Proteomics 2009, 9, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, S. Routine Procedure for Growing Rice Plants in Culture Solution. In Laboratory Manual for Physiological Studies of Rice; Yoshida, S., Forno, D.A., Cock, J.H., Eds.; International Rice Research Institute: Los Banos, Philippines, 1976; pp. 61–66. [Google Scholar]
- Wu, C.; Wang, Q.; Xue, S.; Pan, W.; Lou, L.; Li, D.; Hartley, W. Do aeration conditions affect arsenic and phosphate accumulation and phosphate transporter expression in rice (Oryza sativa L.)? Environ. Sci. Pollut. Res. 2016, 25, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, G.; Pandey, R.N.; Sahoo, R.N.; Gupta, V.K.; Datta, S.C.; Kumar, D. Monitoring nitrogen, phosphorus and sulphur in hybrid rice (Oryza sativa L.) using hyperspectral remote sensing. Precis. Agric. 2016, 18, 736–761. [Google Scholar] [CrossRef]
- Syers, J.K.; Johnston, A.E.; Curtin, D. Efficiency of soil and fertilizer phosphorus use. FAO Fertil. Plant Nutr. Bull. 2008, 18, 108. [Google Scholar]
- Isaacson, T.; Damasceno, C.M.B.; Saravanan, R.S.; He, Y.; Catalá, C.; Saladié, M.; Rose, J.K.C. Sample extraction techniques for enhanced proteomic analysis of plant tissues. Nat. Protoc. 2006, 1, 769–774. [Google Scholar] [CrossRef]
- O’Farrell, P.H. High resolution two-dimensional electrophoresis of proteins. J. Boil. Chem. 1975, 250, 4007–4021. [Google Scholar]
- Wheeler, D.L.; Barrett, T.; Benson, D.A.; Bryant, S.H.; Canese, K.; Chetvernin, V.; Church, D.M.; DiCuccio, M.; Edgar, R.; Federhen, S.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2006, 35, 5–12. [Google Scholar] [CrossRef]
- Wu, C.H. The Universal Protein Resource (UniProt): An expanding universe of protein information. Nucleic Acids Res. 2006, 34, D187–D191. [Google Scholar] [CrossRef]
- Guda, C. pTARGET: A web server for predicting protein subcellular localization. Nucleic Acids Res. 2006, 34, W210–W213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chankaew, S.; Monkham, T.; Pinta, W.; Sanitchon, J.; Kaewpradit, W.; Srinives, P. Screening tolerance to phosphorus deficiency and validation of phosphorus pptake 1 (Pup1) gene linked markers in Thai indigenous upland rice germplasm. Agronomy 2019, 9, 81. [Google Scholar] [CrossRef] [Green Version]
- Wissuwa, M.; Ae, N. Genotypic variation for tolerance to phosphorus deficiency in rice and the potential for its exploitation in rice improvement. Plant Breed. 2001, 120, 43–48. [Google Scholar] [CrossRef]
- Vejchasarn, P.; Lynch, J.P.; Brown, K.M. Genetic Variability in Phosphorus Responses of Rice Root Phenotypes. Rice 2016, 9, 29. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Liu, H.; Tao, P.; Chen, H. Comparative Proteomic Analyses Provide New Insights into Low Phosphorus Stress Responses in Maize Leaves. PLoS ONE 2014, 9, e98215. [Google Scholar] [CrossRef]
- Zhang, W.; Gruszewski, H.A.; Chevone, B.I.; Nessler, C.L. An Arabidopsis Purple Acid Phosphatase with Phytase Activity Increases Foliar Ascorbate1 [OA]. Plant Physiol. 2007, 146, 431–440. [Google Scholar] [CrossRef] [Green Version]
- Bar-Even, A. Daring metabolic designs for enhanced plant carbon fixation. Plant Sci. 2018, 273, 71–83. [Google Scholar] [CrossRef] [PubMed]
- De Porcellinis, A.J.; Nørgaard, H.; Brey, L.M.F.; Erstad, S.M.; Jones, P.R.; Heazlewood, J.L.; Sakuragi, Y. Overexpression of bifunctional fructose-1,6-bisphosphatase/sedoheptulose-1,7-bisphosphatase leads to enhanced photosynthesis and global reprogramming of carbon metabolism in Synechococcus sp. PCC 7002. Metab. Eng. 2018, 47, 170–183. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lu, Y.; Chang, Z.; Wang, S.; Ding, Y.; Ding, C. Transcriptomic analysis of field-grown rice (Oryza sativa L.) reveals responses to shade stress in reproductive stage. Plant Growth Regul. 2018, 84, 583–592. [Google Scholar] [CrossRef]
- Huang, S.; Millar, A.H. Succinate dehydrogenase: The complex roles of a simple enzyme. Curr. Opin. Plant Boil. 2013, 16, 344–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belt, K.; Van Aken, O.; Murcha, M.; Millar, A.H.; Huang, S. An Assembly Factor Promotes Assembly of Flavinated SDH1 into the Succinate Dehydrogenase Complex. Plant Physiol. 2018, 177, 1439–1452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheibe, R. Maintaining homeostasis by controlled alternatives for energy distribution in plant cells under changing conditions of supply and demand. Photosynth. Res. 2018, 139, 81–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Troncoso-Ponce, M.; Rivoal, J.; Dorion, S.; Sánchez, R.; Venegas-Calerón, M.; Moreno-Pérez, A.J.; Baud, S.; Garces, R.; Martínez-Force, E. Molecular and biochemical characterization of the sunflower (Helianthus annuus L.) cytosolic and plastidial enolases in relation to seed development. Plant Sci. 2018, 272, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Yi, K.; Wu, Z.; Zhou, J.; Du, L.; Guo, L.; Wu, Y.; Wu, P. OsPTF1, a Novel Transcription Factor Involved in Tolerance to Phosphate Starvation in Rice1 [w]. Plant Physiol. 2005, 138, 2087–2096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, F.; Li, Q.; Sun, L.; He, Z. The Rice 14-3-3 Gene Family and its Involvement in Responses to Biotic and Abiotic Stress. DNA Res. 2006, 13, 53–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zörb, C.; Ludewig, U.; Hawkesford, M.J. Perspective on Wheat Yield and Quality with Reduced Nitrogen Supply. Trends Plant Sci. 2018, 23, 1029–1037. [Google Scholar] [CrossRef] [Green Version]
- Kudlicki, W.; Coffman, A.; Kramer, G.; Hardesty, B. Renaturation of rhodanese by translational elongation factor (EF) Tu. Protein refolding by EF-Tu flexing. J. Boil. Chem. 1997, 272, 32206–32210. [Google Scholar] [CrossRef] [Green Version]
- Yakobov, N.; Debard, S.; Fischer, F.; Senger, B.; Becker, H. Cytosolic aminoacyl-tRNA synthetases: Unanticipated relocations for unexpected functions. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2018, 1861, 387–400. [Google Scholar] [CrossRef]
- Hummel, M.; Cordewener, J.H.; de Groot, J.C.; Smeekens, S.; America, A.H.; Hanson, J. Dynamic protein composition of Arabidopsis thaliana cytosolic ribosomes in response to sucrose feeding as revealed by label free MS E proteomics. Proteomics 2012, 12, 1024–1038. [Google Scholar] [CrossRef]
- Graifer, D.M.; Karpova, G. Roles of ribosomal proteins in the functioning of translational machinery of eukaryotes. Biochimie 2015, 109, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Cardona, T.; Shao, S.; Nixon, P.J. Enhancing photosynthesis in plants: The light reactions. Essays Biochem. 2018, 62, 85–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalal, V.K.; Tripathy, B.C. Water-stress induced downsizing of light-harvesting antenna complex protects developing rice seedlings from photo-oxidative damage. Sci. Rep. 2018, 8, 5955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashida, S.-N.; Miyagi, A.; Nishiyama, M.; Yoshida, K.; Hisabori, T.; Kawai-Yamada, M. Ferredoxin/thioredoxin system plays an important role in the chloroplastic NADP status of Arabidopsis. Plant J. 2018, 95, 947–960. [Google Scholar] [CrossRef] [PubMed]
- Medrano, H.; Parry, M.A.J.; Socias, X.; Lawlor, D.W. Long term water stress inactivates Rubisco in subterranean clover. Ann. Appl. Boil. 1997, 131, 491–501. [Google Scholar] [CrossRef]
- Yousuf, P.Y.; Abd Allah, E.F.; Nauman, M.; Asif, A.; Hashem, A.; Alqarawi, A.A.; Ahmad, A. Responsive Proteins in Wheat Cultivars with Contrasting Nitrogen Efficiencies under the Combined Stress of High Temperature and Low Nitrogen. Genes 2017, 8, 356. [Google Scholar] [CrossRef] [Green Version]
- Henkes, S.; Sonnewald, U.; Badur, R.; Flachmann, R.; Stitt, M. A small decrease of plastid transketolase activity in antisense tobacco transformants has dramatic effects on photosynthesis and phenylpropanoid metabolism. Plant Cell 2001, 13, 535–551. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.-W.; Rakwal, R.; Agrawal, G.K.; Jung, Y.-H.; Shibato, J.; Jwa, N.-S.; Iwahashi, Y.; Iwahashi, H.; Kim, D.H.; Shim, I.-S.; et al. A hydroponic rice seedling culture model system for investigating proteome of salt stress in rice leaf. Electrophoresis 2005, 26, 4521–4539. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Liu, D.; Tschaplinski, T.J.; Tuskan, G.A. Comparative genomics can provide new insights into the evolutionary mechanisms and gene function in CAM plants. J. Exp. Bot. 2019, 70, 6539–6547. [Google Scholar] [CrossRef]
- Liu, X.; Dai, C.; Zhou, J.; Ren, C.; Li, X.; Zhang, C.; Zhang, J. Phosphoenolpyruvate carboxylase regulation in C4-PEPC -expressing transgenic rice during early responses to drought stress. Physiol. Plant. 2016, 159, 178–200. [Google Scholar] [CrossRef]
- Zhang, Y.; Giuliani, R.; Zhang, Y.; Zhang, Y.; Araújo, W.L.; Wang, B.; Liu, P.; Sun, Q.; Cousins, A.; Edwards, G.; et al. Characterization of maize leaf pyruvate orthophosphate dikinase using high throughput sequencing. J. Integr. Plant Boil. 2018, 60, 670–690. [Google Scholar] [CrossRef]
- Liu, F.; Huang, N.; Wang, L.; Ling, H.; Sun, T.; Ahmad, W.; Muhammad, K.; Guo, J.; Xu, L.; Gao, S.; et al. A Novel L-ascorbate Peroxidase 6 Gene, ScAPX6, Plays an Important Role in the Regulation of Response to Biotic and Abiotic Stresses in Sugarcane. Front. Plant Sci. 2018, 8, 2262. [Google Scholar] [CrossRef] [Green Version]
- Tao, J.; Wu, H.; Li, Z.; Huang, C.; Xu, X. Molecular Evolution of GDP-D-Mannose Epimerase (GME), a Key Gene in Plant Ascorbic Acid Biosynthesis. Front. Plant Sci. 2018, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Maguire, M.; Coates, A.R.M.; Henderson, B. Chaperonin 60 unfolds its secrets of cellular communication. Cell Stress Chaperones 2002, 7, 317–329. [Google Scholar] [CrossRef] [Green Version]
- Alavilli, H.; Lee, H.; Park, M.; Yun, D.-J.; Lee, B.-H. Enhanced multiple stress tolerance in Arabidopsis by overexpression of the polar moss peptidyl prolyl isomerase FKBP12 gene. Plant Cell Rep. 2017, 37, 453–465. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Kumar, D.; Jain, M.; Chaudhary, P.; Deswal, R.; Sarin, N.B. Ectopic overexpression of a salt stress-induced pathogenesis-related class 10 protein (PR10) gene from peanut (Arachis hypogaea L.) affords broad spectrum abiotic stress tolerance in transgenic tobacco. Plant Cell Tissue Organ Cult. (PCTOC) 2011, 109, 19–31. [Google Scholar] [CrossRef]




| Physiological Traits | Cv. Nagina 22 | Cv. Panvel | Statistical Analysis (p < 0.05) | ||||
|---|---|---|---|---|---|---|---|
| Optimum-P | Low-P | Optimum-P | Low-P | C | T | C × T | |
| Root length (cm) | 14.2 ± 2.55 | 18.5 ± 2.78 ** | 12.8 ± 1.98 | 15.3 ± 1.67 * | 0.031 | 0.012 | 0.033 |
| Shoot length (cm) | 32.5 ± 3.66 | 24.3 ± 3.11 * | 36.4 ± 4.23 | 31.6 ± 0.34 * | 0.033 | 0.021 | 0.035 |
| Plant dry weight (g plant−1) | 2.04 ± 0.47 | 1.24 ± 0.35 * | 2.59 ± 0.61 | 2.14 ± 0.37 * | 0.007 | 0.015 | 0.026 |
| Leaf P concentration (mg g−1 DW) | 1.63 ± 0.24 | 1.04 ± 0.21 *** | 2.05 ± 0.32 | 1.57 ± 0.22 ** | 0.013 | 0.003 | 0.007 |
| Root P concentration (mg g−1 DW) | 1.78 ± 0.23 | 1.22 ± 0.20 ** | 2.14 ± 0.33 | 1.72 ± 0.25 * | 0.021 | 0.005 | 0.011 |
| Phosphorus use efficiency (%) | 12.96 ± 1.24 | 8.61 ± 1.03 ** | 15.97 ± 2.01 | 12.54 ± 1.73 * | 0.002 | 0.000 | 0.006 |
| Distribution of DEPs | Cv. Panvel | Cv. Nagina 22 | Cv. Panvel + Cv. Nagina 22 | Total DEPs |
|---|---|---|---|---|
| Up regulated | 21 | 7 | 14 | 42 |
| Down regulated | 4 | 10 | 7 | 21 |
| S. N. | Accession No. | Name of Protein | Exp. MW (kDa) | Exp. Pi | M.S. | No. of Matched Peptides | Location | Process | Mode of Regulation | Relative spot intensity (Optimum P:Low P) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Nagina 22 | Panvel | ||||||||||
| 1 | ABL74560 | Fructose-bisphosphate aldolase | 45.6 | 6.45 | 82 | 12 | Cytosol | Energy | Upregulated | 1.00:3.13 | 1.00:2.91 |
| 2 | XP_015646992 | Succinate dehydrogenase flavoprotein subunit | 65.8 | 6.12 | 134 | 15 | Mitochondrion | Energy | Upregulated | 1.00:3.22 | 1.00:3.50 |
| 3 | BAA00147 | Ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 54.1 | 6.31 | 348 | 15 | Chloroplast | Photosynthesis | Downregulated | 1.00: −3.51 | 1.00: −3.23 |
| 4 | AAS93256 | Glutathione S-transferase | 34.2 | 5.49 | 94 | 11 | Cytoplasm | Oxidative stress | Upregulated | 1.00:2.40 | 1.00:2.91 |
| 5 | XP_015643741 | Enolase 1 | 57.8 | 5.59 | 107 | 11 | Chloroplast | Cell rescue/defense | Upregulated | 1.00:3.00 | 1.00:3.02 |
| 6 | BAT16624 | NADP-dependant malate dehydrogenase | 49.3 | 6.14 | 114 | 13 | Mitochondrion, chloroplast, cytosol | Metabolism | Downregulated | 1.00: −3.32 | 1.00: −3.10 |
| 7 | 2002393A | Photosystem II oxygen-evolving complex protein 1 | 37.2 | 6.87 | 159 | 13 | Chloroplast | Photosynthesis | Upregulated | 1.00:2.54 | 1.00:3.00 |
| 8 | BAV53208 | Small ribosomal protein 4 | 53.2 | 6.93 | 89 | 9 | Ribosome, mitochondrion | Protein synthesis | Downregulated | 1.00: −3.43 | 1.00: −3.11 |
| 9 | AEP20544 | Ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit | 53.3 | 6.19 | 96 | 11 | Chloroplast | Photosynthesis | Downregulated | 1.00: −4.35 | 1.00: −3.72 |
| 10 | AAP12937 | Transposon protein, putative, CACTA, En/Spmsub-class | 56.2 | 6.8 | 86 | 16 | Nucleus | - | Upregulated | 1.00:2.97 | 1.00:3.68 |
| 11 | Q948T6 | Glyoxylase I 7 | 33.4 | 6.3 | 86 | 14 | Peroxisome | Oxidative stress | Upregulated | 1.00:4.20 | 1.00:5.20 |
| 12 | ABR25753 | Chaperonin GroEL | 65.3 | 4.91 | 87 | 10 | Cytoplasm | Protein fate/stabilization | Upregulated | 1.00:3.53 | 1.00:5.11 |
| 13 | AAM74563 | Elongation factor Tu | 52.1 | 4.98 | 123 | 16 | Cytosol, plastid, mitochondrion | Protein synthesis | Downregulated | 1.00: −2.33 | 1.00: −2.72 |
| 14 | NP_973937 | DNA binding transcription factors | 27.8 | 5.7 | 96 | 8 | Nucleus | Transcription/STMs | Upregulated | 1.00:3.22 | 1.00:4.61 |
| 15 | Q2QLY4 | Methionine synthase | 55.1 | 6.5 | 112 | 10 | Cytosol | Metabolism | Upregulated | 1.00:4.21 | 1.00:4.52 |
| 16 | CAD41255 | OSJNBa0067K08.7 | 33.4 | 5.33 | 141 | 8 | - | - | Downregulated | 1.00: −3.84 | 1.00: −3.51 |
| 17 | D7LVK3 | ABA-responsive element binding protein 3 | 32.8 | 5.93 | 123 | 11 | Nucleus | Transcription/STMs | Upregulated | 1.00:4.42 | 1.00:4.61 |
| 18 | P69667 | Ribosomal protein L23 | 10.13 | 7.02 | 81 | 8 | Mitochondrion | Protein synthesis | Downregulated | 1.00: −2.11 | 1.00: −2.02 |
| 19 | Q6NPS8 | NADPH-dependent FMN reductase | 32.1 | 6.55 | 82 | 8 | Nucleus, cytoplasm | Cell cycle/transport | Upregulated | 1.00:3.62 | 1.00:3.64 |
| 20 | AAX85991 | Protein disulfide isomerase | 57.1 | 5.3 | 152 | 6 | Endoplasmic reticulum | Protein fate/stabilization | Upregulated | 1.00:2.71 | 1.00:3.22 |
| 21 | ANG44638 | Maturase K | 20.9 | 6.36 | 87 | 7 | Nucleus | Cell rescue/defense | Upregulated | 1.00:4.84 | 1.00:4.80 |
| S. N. | Accession No. | Name of Protein | Exp. Mw (kDa) | Exp. Pi | M. S. | No. of Matched Peptides | Location | Process | Mode of Regulation | Relative Spot Intensity (Optimum P: Low P) |
|---|---|---|---|---|---|---|---|---|---|---|
| 22 | ACA50522 | 14¨C3¨C3 protein | 34.2 | 4.93 | 112 | 9 | Nucleus | Transcription/STMs | Upregulated | 1.00:3.60 |
| 23 | BAD07865 | Phosphoribulokinase | 46.1 | 5.45 | 91 | 14 | Chloroplast, cytosol | Photosynthesis | Upregulated | 1.00:3.22 |
| 24 | Q0DYB1 | Inorganic pyrophosphatase family protein | 33.2 | 6.1 | 110 | 12 | Cytoplasm | Metabolism | Upregulated | 1.00:4.63 |
| 25 | BAD67774 | Phosphogluconate dehydrogenase | 54.2 | 5.89 | 104 | 14 | Cytoplasm | Metabolism | Upregulated | 1.00:2.92 |
| 26 | BAF92702 | Chaperonin 60 β precursor | 55.1 | 5.4 | 128 | 8 | Mitochondrion | Protein fate/stabilization | Upregulated | 1.00:2.51 |
| 27 | AAB33001 | Sedoheptulose-1,7-bisphosphatase | 41.9 | 5.81 | 143 | 14 | Chloroplast | Metabolism | Downregulated | 1.00: −2.11 |
| 28 | 3E5R_A | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic precursor | 55.2 | 6.47 | 94 | 13 | Chloroplast | Energy | Upregulated | 1.00:3.52 |
| 29 | ABA18619 | 59-epimerase | 43.5 | 5.89 | 104 | 8 | Cytosol | Oxidative stress | Upregulated | 1.00:3.23 |
| 30 | BAG24017 | RNA-binding protein | 49.7 | 5.30 | 106 | 9 | Cytosol, nucleus | Transcription/STMs | Upregulated | 1.00:4.50 |
| 31 | M1NZ56 | Pyrroline-5-carboxylate synthetase | 28.1 | 6.7 | 114 | 13 | Cytoplasm | Cell rescue/defense | Upregulated | 1.00:3.11 |
| 32 | ACJ54888 | Heat shock protein 40 | 28.7 | 5.8 | 96 | 8 | Nucleus, mitochondrion, ER | Protein fate/stabilization | Upregulated | 1.00:3.50 |
| 33 | AAF85973 | PR-10b protein | 17.2 | 6.7 | 23 | 9 | Nucleus | - | Upregulated | 1.00:2.55 |
| 34 | ABA39947 | UDP-glucose epimerase | 42.5 | 5.78 | 102 | 8 | Cytosol | Oxidative stress | Upregulated | 1.00:1.63 |
| 35 | BAS78758 | Os02g0493300 | 56.4 | 6.15 | 87 | 8 | - | - | Upregulated | 1.00:1.80 |
| 36 | AJB98433 | 6-Phosphogluconolactonase | 34.2 | 5.46 | 87 | 10 | Cytosol | Metabolism | Upregulated | 1.00:3.41 |
| 37 | Q40693 | Heat shock protein 70 | 27.5 | 5.3 | 92 | 9 | Nucleus, mitochondrion | Protein fate/stabilization | Upregulated | 1.00:1.50 |
| 38 | BAA11351 | Ribosomal protein S19 | 12.6 | 6.8 | 58 | 13 | Ribosome, mitochondrion | Protein synthesis | Downregulated | 1.00: −3.22 |
| 39 | ABI74568 | Phosphoglycerate kinase | 46.2 | 5.48 | 167 | 14 | Cytosol | Energy | Upregulated | 1.00:3.53 |
| 40 | ABA92415 | NADP-dependent oxidoreductase P1 | 40.3 | 6.31 | 119 | 13 | Chloroplast | Photosynthesis | Upregulated | 1.00:3.28 |
| 41 | AAQ23061 | Heat shock responsive transcription factor | 37.9 | 5.1 | 54 | 9 | Nucleus, cytoplasm | Protein fate/stabilization | Upregulated | 1.00:3.97 |
| 42 | BAC00625 | Malate dehydrogenase | 37.7 | 5.41 | 86 | 9 | Mitochondrion | Metabolism | Downregulated | 1.00: −3.24 |
| 43 | AAK92626 | ATPase | 74.8 | 6.48 | 157 | 11 | Plasma membrane | Energy | Upregulated | 1.00:3.32 |
| 44 | BAD17324 | Flavonol synthase | 35.9 | 5.43 | 104 | 10 | Cytoplasm, nucleus | Oxidative stress | Upregulated | 1.00:4.61 |
| 45 | Q6AVA8 | Pyruvate orthophosphate dikinase | 73.3 | 5.15 | 128 | 22 | Chloroplast, cytoplasm | Metabolism | Downregulated | 1.00: ─3.00 |
| 46 | Q6K9N6 | Succinyl-CoA synthetase beta subunit | 44.2 | 5.64 | 63 | 10 | Mitochondrion | Metabolism | Upregulated | 1.00:3.62 |
| S. N. | Accession No. | Name of Protein | Exp. Mw (kDa) | Exp. Pi | M. S. | No. of Matched Peptides | Location | Process | Mode of Regulation | Relative Spot Intensity (Optimum P: Low P) |
|---|---|---|---|---|---|---|---|---|---|---|
| 47 | P93431 | Rubisco activase chloroplast precursor | 52.21 | 4.93 | 148 | 6 | Chloroplast | Photosynthesis | Downregulated | 1.00: −3.26 |
| 48 | XP_015641702 | Ferredoxin-nitrite reductase, chloroplastic | 71.3 | 6.15 | 94 | 11 | Chloroplast | Photosynthesis | Downregulated | 1.00: −4.43 |
| 49 | BAP76084 | Phosphoenolpyruvate carboxylase | 25.9 | 6.87 | 98 | 7 | Chloroplast, mitochondrion | Metabolism | Downregulated | 1.00: −3.26 |
| 50 | Q01859 | F1-ATP synthase, beta subunit | 66.4 | 5.39 | 165 | 18 | Mitochondrion | Energy | Upregulated | 1.00:4.70 |
| 51 | A3C4S4 | GDP-D-mannose-3′,5′-epimerase | 54.3 | 6.63 | 102 | 11 | Cytosol | Oxidative stress | Upregulated | 1.00:3.32 |
| 52 | ABA96472 | FKBP-type peptidyl-prolyl cis-trans isomerase | 27.2 | 6.84 | 86 | 8 | Nucleus | Cell rescue/defense | Upregulated | 1.00:3.83 |
| 53 | AAS46111 | RNA polymerase β chain | 81.1 | 6.93 | 215 | 10 | Chloroplast | Transcription/STMs | Downregulated | 1.00: −3.90 |
| 54 | AAB65699 | Ferredoxin | 44.5 | 6.35 | 147 | 16 | Chloroplast | Photosynthesis | Downregulated | 1.00: −3.44 |
| 55 | A2Y7D9 | Chloroplast light-harvesting complex I protein LHCA3 | 31.1 | 6.46 | 85 | 7 | Chloroplast | Photosynthesis | Downregulated | 1.00: −3.37 |
| 56 | P48494 | Triosephosphate isomerase | 21.2 | 5.5 | 151 | 13 | Cytoplasm | Metabolism | Downregulated | 1.00: −3.70 |
| 57 | - | Unnamed protein product | 46.3 | 5.49 | 97 | 14 | - | - | Upregulated | 1.00:3.80 |
| 58 | XP_015643207 | Transketolase chloroplastic | 17.8 | 6.12 | 84 | 16 | Chloroplast | Photosynthesis | Downregulated | 1.00: −3.52 |
| 59 | ABL74559 | Glyceraldehyde 3-phosphate dehydrogenase | 53.8 | 6.72 | 84 | 8 | Cytoplasm | Energy | Upregulated | 1.00:3.45 |
| 60 | AET04420 | Integrin-linked protein kinase family protein | 34.2 | 6.11 | 104 | 12 | Ribosome | Transcription/STMs | Downregulated | 1.00: −3.11 |
| 61 | XP_015621402 | Asparaginyl-tRNA synthetase, cytoplasmic 3 | 65.1 | 6.13 | 173 | 23 | Cytoplasm | Protein synthesis | Downregulated | 1.00: −3.24 |
| 62 | AAP13093 | Ascorbate peroxidase cytoplasmic | 31.4 | 5.75 | 92 | 12 | Cytoplasm | Oxidative stress | Upregulated | 1.00:4.42 |
| 63 | AAB63591 | Chaperonin 10 Kd subunit | 25.8 | 5.66 | 82 | 8 | Cytoplasm | Protein stabilization | Upregulated | 1.00:3.54 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tantray, A.Y.; Ali, H.M.; Ahmad, A. Analysis of Proteomic Profile of Contrasting Phosphorus Responsive Rice Cultivars Grown under Phosphorus Deficiency. Agronomy 2020, 10, 1028. https://0-doi-org.brum.beds.ac.uk/10.3390/agronomy10071028
Tantray AY, Ali HM, Ahmad A. Analysis of Proteomic Profile of Contrasting Phosphorus Responsive Rice Cultivars Grown under Phosphorus Deficiency. Agronomy. 2020; 10(7):1028. https://0-doi-org.brum.beds.ac.uk/10.3390/agronomy10071028
Chicago/Turabian StyleTantray, Aadil Yousuf, Hayssam M. Ali, and Altaf Ahmad. 2020. "Analysis of Proteomic Profile of Contrasting Phosphorus Responsive Rice Cultivars Grown under Phosphorus Deficiency" Agronomy 10, no. 7: 1028. https://0-doi-org.brum.beds.ac.uk/10.3390/agronomy10071028