Genetic Diversity and Population Structure of Soybean Lines Adapted to Sub-Saharan Africa Using Single Nucleotide Polymorphism (SNP) Markers
Abstract
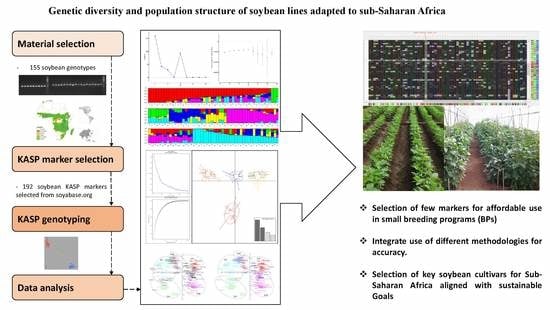
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Sampling
2.2. DNA Extraction and Genotyping with SNP Markers
2.3. Statistical Analysis and Genetic Differentiation of Soybean
3. Results
3.1. Genetic Diversity
3.2. Structure Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hartman, G.L.; West, E.D.; Herman, T.K. Crops that feed the World 2. Soybean—worldwide production, use, and constraints caused by pathogens and pests. Food Secur. 2011, 3, 5–17. [Google Scholar] [CrossRef]
- Foster, R.; Williamson, C.; Lunn, J. BRIEFING PAPER: Culinary oils and their health effects. Nutr. Bull. 2009, 34, 4–47. [Google Scholar] [CrossRef]
- Chander, S.; Ortega-Beltran, A.; Bandyopadhyay, R.; Sheoran, P.; Ige, G.O.; Vasconcelos, M.W.; Garcia-Oliveira, A.L. Prospects for Durable Resistance Against an Old Soybean Enemy: A Four-Decade Journey from Rpp1 (Resistance to Phakopsora pachyrhizi) to Rpp7. Agronomy 2019, 9, 348. [Google Scholar] [CrossRef] [Green Version]
- OECD; FAO. OECD-FAO Agricultural Outlook 2019–2028; OECD Publishing: Paris, France; Food and Agriculture Organization of the United Nations: Rome, Italy, 2019. [Google Scholar] [CrossRef]
- Day, L. Proteins from land plants—Potential resources for human nutrition and food security. Trends Food Sci. Technol. 2013, 32, 25–42. [Google Scholar] [CrossRef]
- Tefera, H.; Kamara, A.Y.; Asafo-Adjei, B.; Dashiell, K.E. Improvement in Grain and Fodder Yields of Early-Maturing Promiscuous Soybean Varieties in the Guinea Savanna of Nigeria. Crop. Sci. 2009, 49, 2037–2042. [Google Scholar] [CrossRef] [Green Version]
- Chikowo, R.; Corbeels, M.; Mapfumo, P.; Tittonell, P.; Vanlauwe, B.; Giller, K.E. Nitrogen and Phosphorus Capture and Recovery Efficiencies, and Crop Responses to a Range of Soil Fertility Management Strategies in Sub-Saharan Africa. In Innovations as Key to the Green Revolution in Africa; Bationo, A., Waswa, B., Okeyo, J., Maina, F., Kihara, J., Eds.; Springer: Dordrecht, The Netherlands, 2011; pp. 571–589. [Google Scholar] [CrossRef]
- Khojely, D.M.; Ibrahim, S.E.; Sapey, E.; Han, T. History, current status, and prospects of soybean production and research in sub-Saharan Africa. Crop. J. 2018, 6, 226–235. [Google Scholar] [CrossRef]
- Mutegi, J.; Zingore, S.I. Boosting Soybean Production for Improved Food Security and Incomes in Africa; Sub-Saharan Africa Program; The International Plant Nutrition Institute (IPNI): Nairobi, Kenya, 2014; Available online: http://ssa.ipni.net/ipniweb/region/africa.nsf/0/28600CA4712A18F685257BE100695F27/$FILE/Soybean%20production%20in%20SSA%20BMPs,%20Challenges%20and%20Opportunities.pdf (accessed on 15 February 2021).
- Giller, K.E.; Dashiell, K.E. Glycine max (L.) Merr. In Plant Resources of Tropical Africa 1. Cereals and Pulses; Brink, M., Belay, G., Eds.; PROTA Foundation: Wageningen, The Netherlands; Backhuys Publishers: Leiden, The Netherlands; CTA: Wageningen, The Netherlands, 2006; pp. 76–82. [Google Scholar]
- Hymowitz, T. The History of the Soybean. In Soybeans: Chemistry, Production, Processing, and Utilization; Johnson, L.A., White, P.J., Galloway, R., Eds.; AOCS Press: Urbana, IL, USA, 2008; pp. 1–31. [Google Scholar]
- Chigeza, G.; Boahen, S.; Gedil, M.; Agoyi, E.; Mushoriwa, H.; Denwar, N.; Gondwe, T.; Tesfaye, A.; Kamara, A.; Alamu, O.E.; et al. Public sector soybean (Glycine max) breeding: Advances in cultivar development in the African tropics. Plant Breed. 2018, 138, 455–464. [Google Scholar] [CrossRef] [Green Version]
- Roldán-Ruiz, I.; van Euwijk, F.; Gilliland, T.J.; Dubreuil, P.; Dillmann, C.; Lallemand, J.; De Loose, M.; Baril, C.P. A comparative study of molecular and morphological methods of describing relationships between perennial ryegrass (Lolium perenne L.) varieties. Theor. Appl. Genet. 2001, 103, 1138–1150. [Google Scholar] [CrossRef]
- Nadeem, M.A.; Nawaz, M.A.; Shahid, M.Q.; Doğan, Y.; Comertpay, G.; Yıldız, M.; Hatipoğlu, R.; Ahmad, F.; Alsaleh, A.; Labhane, N.; et al. DNA molecular markers in plant breeding: Current status and recent advancements in genomic selection and genome editing. Biotechnol. Biotechnol. Equip. 2018, 32, 261–285. [Google Scholar] [CrossRef] [Green Version]
- Semagn, F.K.; Babu, R.; Hearne, S.; Olsen, M. Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): Overview of the technology and its application in crop improvement. Mol. Breed. 2014, 33, 1–14. [Google Scholar] [CrossRef]
- Mammadov, J.; Aggarwal, R.; Buyyarapu, R.; Kumpatla, S. SNP Markers and Their Impact on Plant Breeding. Int. J. Plant Genom. 2012, 2012, 728398. [Google Scholar] [CrossRef] [PubMed]
- Thomson, M.J. High-Throughput SNP Genotyping to Accelerate Crop Improvement. Plant Breed. Biotechnol. 2014, 2, 195–212. [Google Scholar] [CrossRef]
- Ha, B.; Hussey, R.S.; Boerma, H.R. Development of SNP Assays for Marker-Assisted Selection of Two Southern Root-Knot Nematode Resistance QTL in Soybean. Crop. Sci. 2007, 47, S73–S82. [Google Scholar] [CrossRef]
- Adu, G.B.; Badu-Apraku, B.; Akromah, R.; Garcia-Oliveira, A.L.; Awuku, F.J.; Gedil, M. Genetic diversity and population structure of early-maturing tropical maize inbred lines using SNP markers. PLoS ONE 2019, 14, e0214810. [Google Scholar] [CrossRef] [Green Version]
- Saghai-Maroof, M.A.; Soliman, K.M.; Jorgensen, R.A.; Allard, R.W. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. USA 1984, 81, 8014–8018. [Google Scholar] [CrossRef] [Green Version]
- Hyten, D.L.; Choi, I.-Y.; Song, Q.; Specht, J.E.; Carter, T.E.; Shoemaker, R.C.; Hwang, E.-Y.; Matukumalli, L.K.; Cregan, P.B. A High Density Integrated Genetic Linkage Map of Soybean and the Development of a 1536 Universal Soy Linkage Panel for Quantitative Trait Locus Mapping. Crop. Sci. 2010, 50, 960–968. [Google Scholar] [CrossRef] [Green Version]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef] [Green Version]
- Earl, D.A.; Vonholdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2011, 4, 359–361. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [Green Version]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 2010, 11, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perrier, X.; Jacquemoud-Collet, J.P. DARwin Software, 2006.
- Rambaut, A. FigTree-Version 1.4. 3, A Graphical Viewer of Phylogenetic Trees. 2007.
- Chung, G.; Singh, R.J. Broadening the Genetic Base of Soybean: A Multidisciplinary Approach. Crit. Rev. Plant Sci. 2008, 27, 295–341. [Google Scholar] [CrossRef]
- Gupta, S.; Manjaya, J. Genetic diversity and population structure of Indian soybean [Glycine max (L.) Merr.] revealed by simple sequence repeat markers. J. Crop. Sci. Biotechnol. 2017, 20, 221–231. [Google Scholar] [CrossRef]
- Liu, Z.; Li, H.; Wen, Z.; Fan, X.; Li, Y.; Guan, R.; Guo, Y.; Wang, S.; Wang, D.; Qiu, L. Comparison of Genetic Diversity between Chinese and American Soybean (Glycine max (L.)) Accessions Revealed by High-Density SNPs. Front. Plant Sci. 2017, 8, 2014. [Google Scholar] [CrossRef] [Green Version]
- Abe, J.; Xu, D.H.; Suzuki, Y.; Kanazawa, A.; Shimamoto, Y. Soybean germplasm pools in Asia revealed by nuclear SSRs. Theor. Appl. Genet. 2003, 106, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Denwar, N.N.; Awuku, F.J.; Diers, B.; Addae-Frimpomaah, F.; Chigeza, G.; Oteng-Frimpong, R.; Puozaa, D.K.; Barnor, M.T. Genetic diversity, population structure and key phenotypic traits driving variation within soyabean (Glycine max) collection in Ghana. Plant Breed. 2019, 138, 577–587. [Google Scholar] [CrossRef]
- Li, Y.-H.; Li, W.; Zhang, C.; Yang, L.; Chang, R.-Z.; Gaut, B.S.; Qiu, L.-J. Genetic diversity in domesticated soybean (Glycine max) and its wild progenitor (Glycine soja) for simple sequence repeat and single-nucleotide polymorphism loci. New Phytol. 2010, 188, 242–253. [Google Scholar] [CrossRef]
- Semagn, K.; Magorokosho, C.; Vivek, B.S.; Makumbi, D.; Beyene, Y.; Mugo, S.; Prasanna, B.M.; Warburton, M.L. Molecular characterization of diverse CIMMYT maize inbred lines from eastern and southern Africa using single nucleotide polymorphic markers. BMC Genom. 2012, 13, 113. [Google Scholar] [CrossRef] [Green Version]
- Badu-Apraku, B.; Garcia-Oliveira, A.L.; Petroli, C.D.; Hearne, S.; Adewale, S.A.; Gedil, M. Genetic diversity and population structure of early and extra-early maturing maize germplasm adapted to sub-Saharan Africa. BMC Plant Biol. 2021, 21, 96. [Google Scholar] [CrossRef]





| Minor Allele Frequency (MAF) | Heterozygosity | Gene Diversity | Polymorphic Information Content (PIC) | |
|---|---|---|---|---|
| Minimum | 0.043 | 0.000 | 0.082 | 0.079 |
| Median | 0.340 | 0.055 | 0.449 | 0.348 |
| Maximum | 0.500 | 0.190 | 0.500 | 0.375 |
| Mean | 0.328 | 0.058 | 0.414 | 0.324 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chander, S.; Garcia-Oliveira, A.L.; Gedil, M.; Shah, T.; Otusanya, G.O.; Asiedu, R.; Chigeza, G. Genetic Diversity and Population Structure of Soybean Lines Adapted to Sub-Saharan Africa Using Single Nucleotide Polymorphism (SNP) Markers. Agronomy 2021, 11, 604. https://0-doi-org.brum.beds.ac.uk/10.3390/agronomy11030604
Chander S, Garcia-Oliveira AL, Gedil M, Shah T, Otusanya GO, Asiedu R, Chigeza G. Genetic Diversity and Population Structure of Soybean Lines Adapted to Sub-Saharan Africa Using Single Nucleotide Polymorphism (SNP) Markers. Agronomy. 2021; 11(3):604. https://0-doi-org.brum.beds.ac.uk/10.3390/agronomy11030604
Chicago/Turabian StyleChander, Subhash, Ana Luísa Garcia-Oliveira, Melaku Gedil, Trushar Shah, Gbemisola Oluwayemisi Otusanya, Robert Asiedu, and Godfree Chigeza. 2021. "Genetic Diversity and Population Structure of Soybean Lines Adapted to Sub-Saharan Africa Using Single Nucleotide Polymorphism (SNP) Markers" Agronomy 11, no. 3: 604. https://0-doi-org.brum.beds.ac.uk/10.3390/agronomy11030604