Highly Specific Memory B Cells Generation after the 2nd Dose of BNT162b2 Vaccine Compensate for the Decline of Serum Antibodies and Absence of Mucosal IgA
Abstract
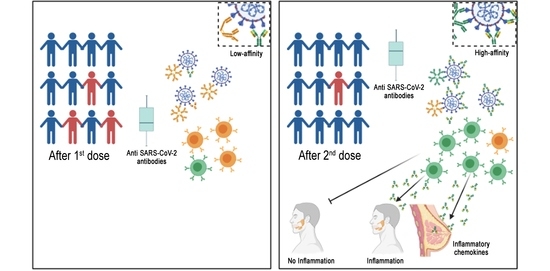
:1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Human Subjects
2.3. Cell Isolation and Cryopreservation
2.4. Polyclonal Memory B-Cell Stimulation
2.5. B Cell ELISpot
2.6. ELISA for Specific IgA and IgM Detection
2.7. Quantitative Determination of Anti-N, Anti-S, Trimeric Spike and RBD Antibodies
2.8. Detection of Antigen-Specific Memory B Cells
2.9. SARS-CoV-2 Variant Determination
2.10. Statistical Analysis
3. Results
3.1. Serum Antibodies
3.2. Memory B Cell Detected by ELISpot
3.3. Spike-Specific Memory B Cells Detected by Flow Cytometry
3.4. IgA Is not Produced at Mucosal Sites after Vaccination with BNT162b2
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data and Materials Availability
Acknowledgments
Conflicts of Interest
References
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 MRNA Covid-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Jackson, L.A.; Anderson, E.J.; Rouphael, N.G.; Roberts, P.C.; Makhene, M.; Coler, R.N.; McCullough, M.P.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; et al. An MRNA Vaccine against SARS-CoV-2—Preliminary Report. N. Engl. J. Med. 2020, 383, 1920–1931. [Google Scholar] [CrossRef]
- Voysey, M.; Clemens, S.A.C.; Madhi, S.A.; Weckx, L.Y.; Folegatti, P.M.; Aley, P.K.; Angus, B.; Baillie, V.L.; Barnabas, S.L.; Bhorat, Q.E.; et al. Safety and Efficacy of the ChAdOx1 NCoV-19 Vaccine (AZD1222) against SARS-CoV-2: An Interim Analysis of Four Randomised Controlled Trials in Brazil, South Africa, and the UK. Lancet 2021, 397, 99–111. [Google Scholar] [CrossRef]
- Mathieu, E.; Ritchie, H.; Ortiz-Ospina, E.; Roser, M.; Hasell, J.; Appel, C.; Giattino, C.; Rodés-Guirao, L. A Global Database of COVID-19 Vaccinations. Nat. Hum. Behav. 2021, 5, 947–953. [Google Scholar] [CrossRef]
- Bieniasz, P. The Case against Delaying SARS-CoV-2 MRNA Vaccine Boosting Doses. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2021. Online ahead of print. [Google Scholar] [CrossRef]
- Saad-Roy, C.M.; Morris, S.E.; Metcalf, C.J.E.; Mina, M.J.; Baker, R.E.; Farrar, J.; Holmes, E.C.; Pybus, O.G.; Graham, A.L.; Levin, S.A.; et al. Epidemiological and Evolutionary Considerations of SARS-CoV-2 Vaccine Dosing Regimes. Science 2021, 372, 363–370. [Google Scholar] [CrossRef]
- Andreano, E.; Rappuoli, R. SARS-CoV-2 Escaped Natural Immunity, Raising Questions about Vaccines and Therapies. Nat. Med. 2021, 27, 759–761. [Google Scholar] [CrossRef]
- Pollard, A.J.; Bijker, E.M. A Guide to Vaccinology: From Basic Principles to New Developments. Nat. Rev. Immunol. 2021, 21, 83–100. [Google Scholar] [CrossRef]
- Yoshida, T.; Mei, H.; Dörner, T.; Hiepe, F.; Radbruch, A.; Fillatreau, S.; Hoyer, B.F. Memory B and Memory Plasma Cells. Immunol. Rev. 2010, 237, 117–139. [Google Scholar] [CrossRef]
- Aranburu, A.; Piano Mortari, E.; Baban, A.; Giorda, E.; Cascioli, S.; Marcellini, V.; Scarsella, M.; Ceccarelli, S.; Corbelli, S.; Cantarutti, N.; et al. Human B-Cell Memory Is Shaped by Age- and Tissue-Specific T-Independent and GC-Dependent Events. Eur J. Immunol. 2017, 47, 327–344. [Google Scholar] [CrossRef]
- Grimsholm, O.; Piano Mortari, E.; Davydov, A.N.; Shugay, M.; Obraztsova, A.S.; Bocci, C.; Marasco, E.; Marcellini, V.; Aranburu, A.; Farroni, C.; et al. The Interplay between CD27dull and CD27bright B Cells Ensures the Flexibility, Stability, and Resilience of Human B Cell Memory. Cell Rep. 2020, 30, 2963–2977. [Google Scholar] [CrossRef] [Green Version]
- Crotty, S. A Brief History of T Cell Help to B Cells. Nat. Rev. Immunol. 2015, 15, 185–189. [Google Scholar] [CrossRef]
- Relative Fluorochrome Brightness. Available online: https://www.bu.edu/flow-cytometry/files/2014/09/Fluorochrome-Chart-Relative-Brightness.pdf (accessed on 17 May 2021).
- Marasco, E.; Farroni, C.; Cascioli, S.; Marcellini, V.; Scarsella, M.; Giorda, E.; Piano Mortari, E.; Leonardi, L.; Scarselli, A.; Valentini, D.; et al. B-Cell Activation with CD40L or CpG Measures the Function of B-Cell Subsets and Identifies Specific Defects in Immunodeficient Patients. Eur. J. Immunol. 2017, 47, 131–143. [Google Scholar] [CrossRef]
- Marcellini, V.; Piano Mortari, E.; Fedele, G.; Gesualdo, F.; Pandolfi, E.; Midulla, F.; Leone, P.; Stefanelli, P.; Tozzi, A.E.; Carsetti, R.; et al. Protection against Pertussis in Humans Correlates to Elevated Serum Antibodies and Memory B Cells. Front. Immunol. 2017, 8, 1158. [Google Scholar] [CrossRef] [Green Version]
- Carsetti, R.; Zaffina, S.; Piano Mortari, E.; Terreri, S.; Corrente, F.; Capponi, C.; Palomba, P.; Mirabella, M.; Cascioli, S.; Palange, P.; et al. Different Innate and Adaptive Immune Responses to SARS-CoV-2 Infection of Asymptomatic, Mild, and Severe Cases. Front. Immunol. 2020, 11, 610300. [Google Scholar] [CrossRef]
- Dan, J.M.; Mateus, J.; Kato, Y.; Hastie, K.M.; Yu, E.D.; Faliti, C.E.; Grifoni, A.; Ramirez, S.I.; Haupt, S.; Frazier, A.; et al. Immunological Memory to SARS-CoV-2 Assessed for up to 8 Months after Infection. Science 2021, 371, eabf4063. [Google Scholar] [CrossRef]
- Ochsenbein, A.F.; Zinkernagel, R.M. Natural Antibodies and Complement Link Innate and Acquired Immunity. Immunology Today 2000, 21, 624–630. [Google Scholar] [CrossRef]
- Selva, K.J.; van de Sandt, C.E.; Lemke, M.M.; Lee, C.Y.; Shoffner, S.K.; Chua, B.Y.; Davis, S.K.; Nguyen, T.H.O.; Rowntree, L.C.; Hensen, L.; et al. Systems Serology Detects Functionally Distinct Coronavirus Antibody Features in Children and Elderly. Nat. Commun. 2021, 12, 2037. [Google Scholar] [CrossRef]
- Throsby, M.; van den Brink, E.; Jongeneelen, M.; Poon, L.L.M.; Alard, P.; Cornelissen, L.; Bakker, A.; Cox, F.; van Deventer, E.; Guan, Y.; et al. Heterosubtypic Neutralizing Monoclonal Antibodies Cross-Protective against H5N1 and H1N1 Recovered from Human IgM+ Memory B Cells. PLoS ONE 2008, 3, e3942. [Google Scholar] [CrossRef] [Green Version]
- Agrati, C.; Castilletti, C.; Goletti, D.; Meschi, S.; Sacchi, A.; Matusali, G.; Bordoni, V.; Petrone, L.; Lapa, D.; Notari, S.; et al. Coordinate Induction of Humoral and Spike Specific T-cell Response in a Cohort of Italian Health Care Workers Receiving Bnt162b2 Mrna Vaccine. Microorganisms 2021, 9, 1315. [Google Scholar] [CrossRef]
- Kohmer, N.; Rühl, C.; Ciesek, S.; Rabenau, H.F. Utility of Different Surrogate Enzyme-Linked Immunosorbent Assays (SELISAs) for Detection of SARS-CoV-2 Neutralizing Antibodies. J. Clin. Med. 2021, 10, 2128. [Google Scholar] [CrossRef]
- Siegrist, C.-A. Vaccine Immunology. In Plotkin’s Vaccine, 7th ed.; Plotkin, S.A., Orenstein, W.A., Offit, P.A., Edwards, K., Eds.; Elsevier: Amsterdam, Netherlands, 2018; pp. 16–34. [Google Scholar]
- Victora, G.D.; Wilson, P.C. Germinal Center Selection and the Antibody Response to Influenza. Cell 2015, 163, 545–548. [Google Scholar] [CrossRef] [Green Version]
- Goldman, A.S.; Chheda, S.; Keeney, S.E.; Schmalstieg, F.C. Immunology of Human Milk and Host Immunity. Fetal Neonatal Physiol. 2011, 1690–1701. [Google Scholar] [CrossRef]
- Ferrari, D.V.D.J.; Polettini, J.; de Moraes, L.L.; de Campos, L.A.; da Silva, M.G.; Saeki, E.K.; Morceli, G. Profile of Pro-Inflammatory Cytokines in Colostrum of Nursing Mothers at the Extremes of Reproductive Age. PLoS ONE 2020, 15, e0231882. [Google Scholar] [CrossRef]
- Michie, C.A.; Tantscher, E.; Schall, T.; Rot, A. Physiological Secretion of Chemokines in Human Breast Milk. Eur. Cytokine Netw. 1998, 9, 123–129. [Google Scholar]
- Brandtzaeg, P. Secretory Immunity with Special Reference to the Oral Cavity. J. Oral Microbiol. 2013, 5. [Google Scholar] [CrossRef]
- Sterlin, D.; Mathian, A.; Miyara, M.; Mohr, A.; Anna, F.; Claër, L.; Quentric, P.; Fadlallah, J.; Devilliers, H.; Ghillani, P.; et al. IgA Dominates the Early Neutralizing Antibody Response to SARS-CoV-2. Sci. Transl. Med. 2020, 13, eabd2223. [Google Scholar] [CrossRef]
- Andreano, E.; Nicastri, E.; Paciello, I.; Pileri, P.; Manganaro, N.; Piccini, G.; Manenti, A.; Pantano, E.; Kabanova, A.; Troisi, M.; et al. Extremely Potent Human Monoclonal Antibodies from COVID-19 Convalescent Patients. Cell 2021, 184, 1821–1835.e16. [Google Scholar] [CrossRef]
- Barnes, C.O.; Jette, C.A.; Abernathy, M.E.; Dam, K.-M.A.; Esswein, S.R.; Gristick, H.B.; Malyutin, A.G.; Sharaf, N.G.; Huey-Tubman, K.E.; Lee, Y.E.; et al. SARS-CoV-2 Neutralizing Antibody Structures Inform Therapeutic Strategies. Nature 2020, 588, 682–687. [Google Scholar] [CrossRef]
- Kumar, N.; Arthur, C.P.; Ciferri, C.; Matsumoto, M.L. Structure of the Secretory Immunoglobulin A Core. Science 2020, 367, eaaz5807. [Google Scholar] [CrossRef]
- Mazanec, M.B.; Coudret, C.L.; Fletcher, D.R. Intracellular Neutralization of Influenza Virus by Immunoglobulin A Anti-Hemagglutinin Monoclonal Antibodies. J. Virol. 1995, 69, 1339–1343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devito, C.; Hinkula, J.; Kaul, R.; Lopalco, L.; Bwayo, J.J.; Plummer, F.; Clerici, M.; Broliden, K. Mucosal and Plasma IgA from HIV-Exposed Seronegative Individuals Neutralize a Primary HIV-1 Isolate. AIDS (Lond. Engl.) 2000, 14, 1917–1920. [Google Scholar] [CrossRef] [PubMed]
- Quinti, I.; Mortari, E.P.; Fernandez Salinas, A.; Milito, C.; Carsetti, R. IgA Antibodies and IgA Deficiency in SARS-CoV-2 Infection. Front. Cell. Infect. Microbiol. 2021, 11, 257. [Google Scholar] [CrossRef]
- Tuaillon, E.; Valea, D.; Becquart, P.; Al Tabaa, Y.; Meda, N.; Bollore, K.; Van de Perre, P.; Vendrell, J.-P. Human Milk-Derived B Cells: A Highly Activated Switched Memory Cell Population Primed to Secrete Antibodies. J. Immunol. 2009, 182, 7155–7162. [Google Scholar] [CrossRef]
- Fox, A.; Marino, J.; Amanat, F.; Krammer, F.; Hahn-Holbrook, J.; Zolla-Pazner, S.; Powell, R.L. Robust and Specific Secretory IgA Against SARS-CoV-2 Detected in Human Milk. iScience 2020, 23, 101735. [Google Scholar] [CrossRef]
- Tosif, S.; Neeland, M.R.; Sutton, P.; Licciardi, P.V.; Sarkar, S.; Selva, K.J.; Do, L.A.H.; Donato, C.; Quan Toh, Z.; Higgins, R.; et al. Immune Responses to SARS-CoV-2 in Three Children of Parents with Symptomatic COVID-19. Nat. Commun. 2020, 11, 5703. [Google Scholar] [CrossRef]
- Ketas, T.J.; Chaturbhuj, D.; Portillo, V.M.C.; Francomano, E.; Golden, E.; Chandrasekhar, S.; Debnath, G.; Díaz-Tapia, R.; Yasmeen, A.; Kramer, K.D.; et al. Antibody Responses to SARS-CoV-2 MRNA Vaccines Are Detectable in Saliva. Pathog. Immun. 2021, 6, 116–134. [Google Scholar] [CrossRef]
- Becker, M.; Dulovic, A.; Junker, D.; Ruetalo, N.; Kaiser, P.D.; Pinilla, Y.T.; Heinzel, C.; Haering, J.; Traenkle, B.; Wagner, T.R.; et al. Immune Response to SARS-CoV-2 Variants of Concern in Vaccinated Individuals. Nat. Commun. 2021, 12, 3109. [Google Scholar] [CrossRef]
- Allie, S.R.; Bradley, J.E.; Mudunuru, U.; Schultz, M.D.; Graf, B.A.; Lund, F.E.; Randall, T.D. The Establishment of Resident Memory B Cells in the Lung Requires Local Antigen Encounter. Nat. Immunol. 2019, 20, 97–108. [Google Scholar] [CrossRef]
- Wang, Z.; Lorenzi, J.C.C.; Muecksch, F.; Finkin, S.; Viant, C.; Gaebler, C.; Cipolla, M.; Hoffmann, H.-H.; Oliveira, T.Y.; Oren, D.A.; et al. Enhanced SARS-CoV-2 Neutralization by Dimeric IgA. Sci. Transl. Med. 2020, 13, eabf1555. [Google Scholar] [CrossRef]
- Levine-Tiefenbrun, M.; Yelin, I.; Katz, R.; Herzel, E.; Golan, Z.; Schreiber, L.; Wolf, T.; Nadler, V.; Ben-Tov, A.; Kuint, J.; et al. Initial Report of Decreased SARS-CoV-2 Viral Load after Inoculation with the BNT162b2 Vaccine. Nat. Med. 2021, 27, 790–792. [Google Scholar] [CrossRef] [PubMed]
- Mudgal, R.; Nehul, S.; Tomar, S. Prospects for Mucosal Vaccine: Shutting the Door on SARS-CoV-2. Hum. Vaccines Immunother. 2020, 16, 2921–2931. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.G.; Burgess, J.L.; Naleway, A.L.; Tyner, H.L.; Yoon, S.K.; Meece, J.; Olsho, L.E.W.; Caban-Martinez, A.J.; Fowlkes, A.; Lutrick, K.; et al. Interim Estimates of Vaccine Effectiveness of BNT162b2 and MRNA-1273 COVID-19 Vaccines in Preventing SARS-CoV-2 Infection Among Health Care Personnel, First Responders, and Other Essential and Frontline Workers–Eight U.S. Locations, December 2020-March. MMWR. Morb. Mortal. Wkly. Rep. 2021, 70, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Mahase, E. Covid-19: Reports from Israel Suggest One Dose of Pfizer Vaccine Could Be Less Effective than Expected. BMJ 2021, 372, n217. [Google Scholar] [CrossRef]
- Takemori, T. B cell Memory and Plasma Cell Development. In Molecular Biology of B Cells, 2nd ed.; Honjo, T., Radbruch, A., Reth, M., Alt, F., Eds.; Elsevier: London, UK, 2015; pp. 227–249. [Google Scholar]





| Study Group | Memory B Cells by ELISpot | Memory B Cells by FACS | Surrogate Neutralization | |
|---|---|---|---|---|
| Number of HCW | 108 | 74 | 34 | 70 |
| Age (mean ± SD) | 46.95 ± 11.35 | 47.04 ± 10.93 | 45.7 ± 11.96 | 48.11 ± 11.45 |
| Sex (M/F) | 31/77 | 17/57 | 14/20 | 17/53 |
| Number of patients | Assay | Total Patients Followed Over Time |
| ELISpot | 74 | |
| FACs | 34 | |
| Surrogate Neutralization | 70 | |
| ELISA | 108 |
| Study Group | |
|---|---|
| Mothers’ age (mean ± SD) | 33.36 ± 4.17 |
| Mothers’ breast milk | 7 not vaccinated 16 vaccinated |
| HCWs Age (Mean ± SD) | 45.6 ± 6.5 |
| Sex | 66.7% F; 33.3% M |
| SARS-CoV-2 infection | 33.3% asymptomatic; 50% paucisymptomatic; 16.7% mild |
| SARS-CoV-2 variant | 66.6% Gamma; 16.7% Alpha; 16.7% n.a. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piano Mortari, E.; Russo, C.; Vinci, M.R.; Terreri, S.; Fernandez Salinas, A.; Piccioni, L.; Alteri, C.; Colagrossi, L.; Coltella, L.; Ranno, S.; et al. Highly Specific Memory B Cells Generation after the 2nd Dose of BNT162b2 Vaccine Compensate for the Decline of Serum Antibodies and Absence of Mucosal IgA. Cells 2021, 10, 2541. https://0-doi-org.brum.beds.ac.uk/10.3390/cells10102541
Piano Mortari E, Russo C, Vinci MR, Terreri S, Fernandez Salinas A, Piccioni L, Alteri C, Colagrossi L, Coltella L, Ranno S, et al. Highly Specific Memory B Cells Generation after the 2nd Dose of BNT162b2 Vaccine Compensate for the Decline of Serum Antibodies and Absence of Mucosal IgA. Cells. 2021; 10(10):2541. https://0-doi-org.brum.beds.ac.uk/10.3390/cells10102541
Chicago/Turabian StylePiano Mortari, Eva, Cristina Russo, Maria Rosaria Vinci, Sara Terreri, Ane Fernandez Salinas, Livia Piccioni, Claudia Alteri, Luna Colagrossi, Luana Coltella, Stefania Ranno, and et al. 2021. "Highly Specific Memory B Cells Generation after the 2nd Dose of BNT162b2 Vaccine Compensate for the Decline of Serum Antibodies and Absence of Mucosal IgA" Cells 10, no. 10: 2541. https://0-doi-org.brum.beds.ac.uk/10.3390/cells10102541