Genetic and Transcription Profile Analysis of Tissue-Specific Anthocyanin Pigmentation in Carrot Root Phloem
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Genetic Mapping of Anthocyanin Pigmentation in the Root Outer and Inner Phloem
2.3. RNA Sequencing and Transcriptome Analysis
2.4. Search of Candidate Genes Conditioning Anthocyanin Pigmentation in the Root OP and IP
2.5. Phylogenetic Analysis
2.6. Real-Time Quantitative Reverse Transcriptase PCR
2.7. Anthocyanin Quantification
3. Results
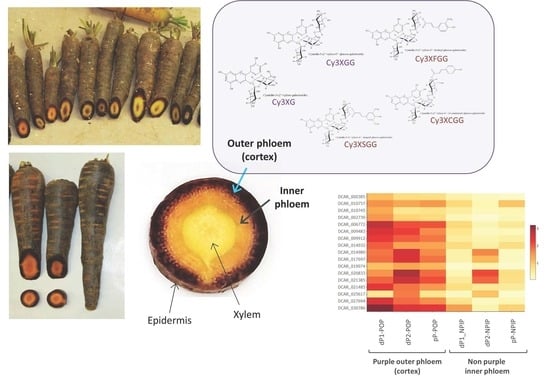
3.1. Genetic Mapping of Phenotypic Pigment Traits
3.2. Transcriptome Comparisons
3.2.1. Differentially Expressed Genes in the ROPAP and RIPAP Regions
3.2.2. Genome-Wide Search of DEGs Associated with Anthocyanin Pigmentation in the Root Outer and Inner Phloem
Regulatory Genes
Structural Anthocyanin Biosynthetic Genes
Genes Involved in Intracellular Transport of Anthocyanins
3.3. Preliminary Identification of Candidate Genes Based on Linkage, Transcriptome, and Phylogenetic Analyses
3.4. Expression Analysis by RT-qPCR of Selected Candidate Genes in 3242 and B7262 Genetic Backgrounds
3.5. RT-qPCR Analysis of Major Candidate Genes for ROPAP and RIPAP in Fully-Pigmented Carrot Roots
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Koes:, R.; Quattrocchio, F.; Mol, J.N.M. The flavonoid biosynthetic pathway in plants: Function and evolution. BioEssays 1994, 16, 123–132. [Google Scholar] [CrossRef]
- Shirley, B.W. Flavonoid biosynthesis: “New” functions for an “old” pathway. Trends Plant Sci. 1996, 1, 377–382. [Google Scholar] [CrossRef]
- Prior, R.L.; Wu, X. Anthocyanins: Structural characteristics that result in unique metabolic patterns and biological activities. Free Radic. Res. 2006, 40, 1014–1028. [Google Scholar] [CrossRef]
- He, J.; Giusti, M.M. Anthocyanins: Natural Colorants with Health-Promoting Properties. Annu. Rev. Food Sci. Technol. 2010, 1, 163–187. [Google Scholar] [CrossRef]
- Koes, R.; Verweij, W.; Quattrocchio, F. Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005, 10, 236–242. [Google Scholar] [CrossRef]
- Simon, P.W. Inheritance and expression of purple and yellow storage root color in carrot. J. Hered. 1996, 87, 63–66. [Google Scholar] [CrossRef] [Green Version]
- Vivek, B.S.; Simon, P.W. Linkage relationships among molecular markers and storage root traits of carrot (Daucus carota L. ssp. sativus). Theor. Appl. Genet. 1999, 99, 58–64. [Google Scholar] [CrossRef]
- Yildiz, M.; Willis, D.K.; Cavagnaro, P.F.; Iorizzo, M.; Abak, K.; Simon, P.W. Expression and mapping of anthocyanin biosynthesis genes in carrot. Theor. Appl. Genet. 2013, 126, 1689–1702. [Google Scholar] [CrossRef]
- Cavagnaro, P.F.; Iorizzo, M.; Yildiz, M.; Senalik, D.; Parsons, J.; Ellison, S.; Simon, P.W. A gene-derived SNP-based high resolution linkage map of carrot including the location of QTL conditioning root and leaf anthocyanin pigmentation. BMC Genom. 2014, 15, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Iorizzo, M.; Curaba, J.; Pottorff, M.; Ferruzzi, M.G.; Simon, P.W.; Cavagnaro, P.F. Carrot anthocyanins genetics and genomics: Status and perspectives to improve its application for the food colorant industry. Genes 2020, 11, 906. [Google Scholar] [CrossRef]
- Iorizzo, M.; Cavagnaro, P.; Bostan, H.; Zhao, Y.; Zhang, J.; Simon, P.W. A Cluster of MYB transcription factors regulates anthocyanin biosynthesis in carrot (Daucus carota L.) root and petiole. Front. Plant Sci. 2019, 9, 1927. [Google Scholar] [CrossRef] [Green Version]
- Bannoud, F.; Ellison, S.; Paolinelli, M.; Horejsi, T.; Senalik, D.; Fanzone, M.; Iorizzo, M.; Simon, P.W.; Cavagnaro, P.F. Dissecting the genetic control of root and leaf tissue-specific anthocyanin pigmentation in carrot (Daucus carota L.). Theor. Appl. Genet. 2019, 132, 2485–2507. [Google Scholar] [CrossRef]
- Xu, Z.S.; Yang, Q.Q.; Feng, K.; Xiong, A.S. Changing carrot color: Insertions in DcMYB7 alter the regulation of anthocyanin biosynthesis and modification. Plant Physiol. 2019, 181, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.S.; Yang, Q.Q.; Feng, K.; Yu, X.; Xiong, A.S. DcMYB113, a root-specific R2R3-MYB, conditions anthocyanin biosynthesis and modification in carrot. Plant Biotechnol. J. 2020, 18, 1585–1597. [Google Scholar] [CrossRef] [Green Version]
- Kodama, M.; Brinch-Pedersen, H.; Sharma, S.; Holme, I.B.; Joernsgaard, B.; Dzhanfezova, T.; Amby, D.B.; Vieira, F.G.; Liu, S.; Gilbert, M.T.P. Identification of transcription factor genes involved in anthocyanin biosynthesis in carrot (Daucus carota L.) using RNA-Seq. BMC Genom. 2018, 19, 1–13. [Google Scholar] [CrossRef]
- Meng, G.; Clausen, S.K.; Rasmussen, S.K. Transcriptome analysis reveals candidate genes related to anthocyanin biosynthesis in different carrot genotypes and tissues. Plants 2020, 9, 344. [Google Scholar] [CrossRef] [Green Version]
- Curaba, J.; Bostan, H.; Cavagnaro, P.F.; Senalik, D.; Mengist, M.F.; Zhao, Y.; Simon, P.W.; Iorizzo, M. Identification of an SCPL gene controlling anthocyanin acylation in carrot (Daucus carota L.) root. Front. Plant Sci. 2020, 10, 1–17. [Google Scholar] [CrossRef]
- Xu, Z.S.; Ma, J.; Wang, F.; Ma, H.Y.; Wang, Q.X.; Xiong, A.S. Identification and characterization of DcUCGalT1, a galactosyltransferase responsible for anthocyanin galactosylation in purple carrot (Daucus carota L.) taproots. Sci. Rep. 2016, 6, 27356. [Google Scholar] [CrossRef] [Green Version]
- Cavagnaro, P.F.; Iorizzo, M. Carrot Anthocyanin Diversity, Genetics, and Genomics. In The Carrot Genome; Simon, P., Iorizzo, M., Grzebelus, D., Baranski, R., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 261–277. ISBN 978-3-030-03389-7. [Google Scholar]
- Broman, K.W.; Wu, H.; Sen, Ś.; Churchill, G.A. R/qtl: QTL mapping in experimental crosses. Bioinformatics 2003, 19, 889–890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Ooijen, J.W. JoinMap® 4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations; Kyazma B.V.: Wageningen, The Netherlands, 2006. [Google Scholar]
- Iorizzo, M.; Ellison, S.; Senalik, D.; Zeng, P.; Satapoomin, P.; Huang, J.; Bowman, M.; Iovene, M.; Sanseverino, W.; Cavagnaro, P.; et al. A high-quality carrot genome assembly provides new insights into carotenoid accumulation and asterid genome evolution. Nat. Genet. 2016, 48, 657–666. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [Green Version]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef] [Green Version]
- Pires, N.; Dolan, L. Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 2010, 27, 862–874. [Google Scholar] [CrossRef] [Green Version]
- Feller, A.; MacHemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amato, A.; Cavallini, E.; Zenoni, S.; Finezzo, L.; Begheldo, M.; Ruperti, B.; Tornielli, G.B. A grapevine TTG2-like WRKY transcription factor is involved in regulating vacuolar transport and flavonoid biosynthesis. Front. Plant Sci. 2017, 7, 1–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rio, D.C.; Ares, M.; Hannon, G.J.; Nilsen, T.W. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb. Protoc. 2010, 2010, pdb.prot5439. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.H.; Karlen, Y.; Bakker, O.; van den Hoff, M.J.B.; Moorman, A.F.M. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef] [Green Version]
- Ramakers, C.; Ruijter, J.M.; Deprez, R.H.L.; Moorman, A.F.M. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef]
- Di Rienzo, J. fgStatistics. Statistical Software for the Analysis of Experiments of Functional Genomics. 2012. Available online: http://sites.google.com/site/fgStatistics/ (accessed on 16 February 2021).
- Tian, C.; Jiang, Q.; Wang, F.; Wang, G.L.; Xu, Z.S.; Xiong, A.S. Selection of suitable reference genes for qPCR normalization under abiotic stresses and hormone stimuli in carrot leaves. PLoS ONE 2015, 10, e0117569. [Google Scholar] [CrossRef] [Green Version]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3-new capabilities and interfaces. Nucleic. Acids. Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thornton, B.; Basu, C. Real-time PCR (qPCR) primer design using free online software. Biochem. Mol. Biol. Educ. 2011, 39, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.S.; Feng, K.; Que, F.; Wang, F.; Xiong, A.S. A MYB transcription factor, DcMYB6, is involved in regulating anthocyanin biosynthesis in purple carrot taproots. Sci. Rep. 2017, 7, 45324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaakola, L.; Poole, M.; Jones, M.O.; Kamarainen-Karppinen, T.; Koskimaki, J.J.; Hohtola, A.; Haggman, H.; Fraser, P.D.; Manning, K.; King, G.J.; et al. A SQUAMOSA MADS Box gene involved in the regulation of anthocyanin accumulation in bilberry fruits. Plant Physiol. 2010, 153, 1619–1629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Behrens, C.E.; Smith, K.E.; Iancu, C.V.; Choe, J.Y.; Dean, J.V. Transport of anthocyanins and other flavonoids by the Arabidopsis ATP-Binding cassette transporter AtABCC2. Sci. Rep. 2019, 9, 437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez, C.; Terrier, N.; Torregrosa, L.; Vialet, S.; Alexandre, F.L.; Verriès, C.; Souquet, J.M.; Mazauric, J.P.; Klein, M.; Cheynier, V.; et al. Grapevine MATE-Type proteins act as vacuolar H+-dependent acylated anthocyanin transporters. Plant Physiol. 2009, 150, 402–415. [Google Scholar] [CrossRef] [Green Version]
- Yao, G.; Ming, M.; Allan, A.C.; Gu, C.; Li, L.; Wu, X.; Wang, R.; Chang, Y.; Qi, K.; Zhang, S.; et al. Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J. 2017, 92, 437–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marrs, K.A. The functions and regulation of glutathione s-transferases in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 127–158. [Google Scholar] [CrossRef]
- Meng, L.; Xu, M.; Wan, W.; Yu, F.; Li, C.; Wang, J.; Wei, Z. Sucrose signaling regulates anthocyanin biosynthesis. Genetics 2018, 210, 607–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esau, K. Developmental anatomy of the fleshy storage organ of Daucus carota. Hilgardia 1940, 13, 175–226. [Google Scholar] [CrossRef] [Green Version]
- Lalusin, A.G.; Nishita, K.; Kim, S.H.; Ohta, M.; Fujimura, T. A new MADS-box gene (IbMADS10) from sweet potato (Ipomoea batatas (L.) Lam) is involved in the accumulation of anthocyanin. Mol. Genet. Genomics 2006, 275, 44–54. [Google Scholar] [CrossRef]
- Wang, R.; Ming, M.; Li, J.; Shi, D.; Qiao, X.; Li, L.; Zhang, S.; Wu, J. Genome-wide identification of the MADS-box transcription factor family in pear (Pyrus bretschneideri) reveals evolution and functional divergence. PeerJ 2017, 5, e3776. [Google Scholar] [CrossRef] [Green Version]
- Nesi, N. The TRANSPARENT TESTA16 locus encodes the ARABIDOPSIS BSISTER MADS domain protein and is required for proper development and pigmentation of the seed coat. Plant Cell Online 2002, 14, 2463–2479. [Google Scholar] [CrossRef] [Green Version]



| Rait | Chr. | Max LOD Position † | Nearest Marker | Position of Nearest Marker (cM) | Max. LOD | 1.5 LOD Interval (nt) | Difference LOD Interval (nt) | 1.5 LOD Interval (cM) | Difference LOD Interval (cM) | % Variance Explained |
|---|---|---|---|---|---|---|---|---|---|---|
| ROPAP | 3 | 3_4002091 | 3_4002091 | 22.9 | 17.4 | 3,394,611–4,075,544 | 680,933 | 21.6–23.5 | 1.9 | 23.6 |
| ROPAP | 3 | Chr3.loc63 | 3_27356688 | 63.2 | 13.9 | 26,273,113–28,568,500 | 2,295,387 | 58.8–65.3 | 6.6 | 17.8 |
| RIPAP | 3 | 3_27356688 | 3_27356688 | 63.2 | 5.3 | 19,453,199–29,895,002 | 10,441,803 | 46.5–67.5 | 21.1 | 14.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bannoud, F.; Carvajal, S.; Ellison, S.; Senalik, D.; Gomez Talquenca, S.; Iorizzo, M.; Simon, P.W.; Cavagnaro, P.F. Genetic and Transcription Profile Analysis of Tissue-Specific Anthocyanin Pigmentation in Carrot Root Phloem. Genes 2021, 12, 1464. https://0-doi-org.brum.beds.ac.uk/10.3390/genes12101464
Bannoud F, Carvajal S, Ellison S, Senalik D, Gomez Talquenca S, Iorizzo M, Simon PW, Cavagnaro PF. Genetic and Transcription Profile Analysis of Tissue-Specific Anthocyanin Pigmentation in Carrot Root Phloem. Genes. 2021; 12(10):1464. https://0-doi-org.brum.beds.ac.uk/10.3390/genes12101464
Chicago/Turabian StyleBannoud, Florencia, Sofia Carvajal, Shelby Ellison, Douglas Senalik, Sebastian Gomez Talquenca, Massimo Iorizzo, Philipp W. Simon, and Pablo F. Cavagnaro. 2021. "Genetic and Transcription Profile Analysis of Tissue-Specific Anthocyanin Pigmentation in Carrot Root Phloem" Genes 12, no. 10: 1464. https://0-doi-org.brum.beds.ac.uk/10.3390/genes12101464