Improved SNV Discovery in Barcode-Stratified scRNA-seq Alignments
Abstract
:1. Introduction
2. Methods
2.1. Sequencing Datasets
2.2. Data Processing: Alignment, Processing, Generation of Individual scRNA-seq Alignments
2.3. Variant Call
2.4. Gene Expression Estimation from scRNA-seq Data
2.5. Cell Type Assessments
2.6. VAFRNA Estimation
2.7. Correlation between VAFRNA and Gene Expression
2.8. Statistical Analyses
3. Results
3.1. Analytical Pipeline
3.2. SNV Calls across TES, WGS, and scRNA-seq
3.3. Transcriptome-Wide SNVs Called Exclusively in the Individual Alignments
3.4. Novel and Known SNVs in the Individual scRNA-seq Alignments
3.5. SceSNVs Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhou, W.; Yang, F.; Xu, Z.; Luo, M.; Wang, P.; Guo, Y.; Nie, H.; Yao, L.; Jiang, Q. Comprehensive Analysis of Copy Number Variations in Kidney Cancer by Single-Cell Exome Sequencing. Front. Genet. 2020, 10, 1379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Dong, X.; Lee, M.; Maslov, A.Y.; Wang, T.; Vijg, J. Single-cell whole-genome sequencing reveals the functional landscape of somatic mutations in B lymphocytes across the human lifespan. Proc. Natl. Acad. Sci. USA 2019, 116, 9014–9019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laks, E.; McPherson, A.; Zahn, H.; Lai, D.; Steif, A.; Brimhall, J.; Biele, J.; Wang, B.; Masud, T.; Ting, J.; et al. Clonal Decomposition and DNA Replication States Defined by Scaled Single-Cell Genome Sequencing. Cell 2019, 179, 1207–1221.e22. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Jiang, Y.; Lam, K.-W.G.; Berletch, J.B.; Disteche, C.M.; Noble, W.S.; Steemers, F.J.; Camerini-Otero, R.D.; Adey, A.C.; Shendure, J. High-Throughput Single-Cell Sequencing with Linear Amplification. Mol. Cell 2019, 76, 676–690.e10. [Google Scholar] [CrossRef]
- Ross, E.; Markowetz, F. OncoNEM: Inferring tumor evolution from single-cell sequencing data. Genome Biol. 2016, 17, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Schnepp, P.M.; Chen, M.; Keller, E.T.; Zhou, X. SNV identification from single-cell RNA sequencing data. Hum. Mol. Genet. 2019, 28, 3569–3583. [Google Scholar] [CrossRef]
- Liu, F.; Zhang, Y.; Zhang, L.; Li, Z.; Fang, Q.; Gao, R.; Zhang, Z. Systematic comparative analysis of single-nucleotide variant detection methods from single-cell RNA sequencing data. Genome Biol. 2019, 20, 242. [Google Scholar] [CrossRef] [Green Version]
- Vu, T.N.; Nguyen, H.-N.; Calza, S.; Kalari, K.R.; Wang, L.; Pawitan, Y. Cell-level somatic mutation detection from single-cell RNA sequencing. Bioinformatics 2019, 35, 4679–4687. [Google Scholar] [CrossRef]
- Petti, A.A.; Williams, S.R.; Miller, C.A.; Fiddes, I.T.; Srivatsan, S.N.; Chen, D.Y.; Fronick, C.C.; Fulton, R.S.; Church, D.M.; Ley, T.J. A general approach for detecting expressed mutations in AML cells using single cell RNA-sequencing. Nat. Commun. 2019, 10, 3660. [Google Scholar] [CrossRef]
- Prashant, N.M.; Liu, H.; Bousounis, P.; Spurr, L.; Alomran, N.; Ibeawuchi, H.; Sein, J.; Reece-Stremtan, D.; Horvath, A. Estimating the Allele-Specific Expression of SNVs From 10× Genomics Single-Cell RNA-Sequencing Data. Genes 2020, 11, 240. [Google Scholar]
- Liu, H.; Prashant, N.M.; Spurr, L.F.; Bousounis, P.; Alomran, N.; Ibeawuchi, H.; Sein, J.; Słowiński, P.; Tsaneva-Atanasova, K.; Horvath, A. scReQTL: An approach to correlate SNVs to gene expression from individual scRNA-seq datasets. BMC Genom. 2021, 22, 40. [Google Scholar] [CrossRef]
- Kaminow, B.; Yunusov, D.; Dobin, A. STARsolo: Accurate, fast and versatile mapping/quantification of single-cell and single-nucleus RNA-seq data. bioRxiv 2021. [Google Scholar] [CrossRef]
- Ding, J.; Lin, C.; Bar-Joseph, Z. Cell lineage inference from SNP and scRNA-Seq data. Nucleic Acids Res. 2019, 10, e56. [Google Scholar] [CrossRef] [Green Version]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From fastQ data to high-confidence variant calls: The genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar]
- Kim, S.; Scheffler, K.; Halpern, A.L.; Bekritsky, M.A.; Noh, E.; Källberg, M.; Chen, X.; Kim, Y.; Beyter, D.; Krusche, P.; et al. Strelka2: Fast and accurate calling of germline and somatic variants. Nat. Methods 2018, 15, 591–594. [Google Scholar] [CrossRef]
- MuTect2-GATK. Available online: https://gatk.broadinstitute.org/hc/en-us/articles/360037593851-Mutect2 (accessed on 5 July 2021).
- Wilson, G.W.; Derouet, M.; Darling, G.E.; Yeung, J.C. scSNV: Accurate dscRNA-seq SNV co-expression analysis using duplicate tag collapsing. Genome Biol. 2021, 22, 1–27. [Google Scholar] [CrossRef]
- Prashant, N.M.; Alomran, N.; Chen, Y.; Liu, H.; Bousounis, P.; Movassagh, M.; Edwards, N.; Horvath, A. SCReadCounts: Estimation of cell-level SNVs from scRNA-seq data. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ben-David, U.; Siranosian, B.; Ha, G.; Tang, H.; Oren, Y.; Hinohara, K.; Strathdee, C.A.; Dempster, J.; Lyons, N.J.; Burns, R.; et al. Genetic and transcriptional evolution alters cancer cell line drug response. Nature 2018, 560, 325–330. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2012, 29, 15–21. [Google Scholar] [CrossRef]
- split_bams_by_barcodes. Available online: https://gist.github.com/winni2k/978b33d62fee5e3484ec757de1a00412 (accessed on 3 May 2021).
- Li, H.; Handsaker, R.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butler, A.; Hoffman, P.; Smibert, P.; Papalexi, E.; Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018, 36, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Hafemeister, C.; Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019, 20, 269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aran, D.; Looney, A.P.; Liu, L.; Wu, E.; Fong, V.; Hsu, A.; Chak, S.; Naikawadi, R.P.; Wolters, P.J.; Abate, A.R.; et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019, 20, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Shabalin, A.A. Matrix eQTL: Ultra fast eQTL analysis via large matrix operations. Bioinformatics. 20Shabalin, A.A. Matrix eQTL: Ultra fast eQTL analysis via large matrix operations. Bioinformatics 2012, 28, 1353–1358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spurr, L.; Alomran, N.; Bousounis, P.; Reece-Stremtan, D.; Prashant, N.M.; Liu, H.; Słowiński, P.; Li, M.; Zhang, Q.; Sein, J.; et al. ReQTL: Identifying correlations between expressed SNVs and gene expression using RNA-sequencing data. Bioinformatics 2019, 36, 1351–1359. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Poirion, O.; Zhu, X.; Ching, T.; Garmire, L.X. Using single nucleotide variations in single-cell RNA-seq to identify subpopulations and genotype-phenotype linkage. Nat. Commun. 2018, 9, 4892. [Google Scholar] [CrossRef]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue of Somatic Mutations In Cancer. Nucleic Acids Res. 2018, 47, D941–D947. [Google Scholar] [CrossRef] [Green Version]
- Picardi, E.; D’Erchia, A.M.; Giudice, C.L.; Pesole, G. REDIportal: A comprehensive database of A-to-I RNA editing events in humans. Nucleic Acids Res. 2016, 45, D750–D757. [Google Scholar] [CrossRef] [Green Version]
- Dou, Y.; Gold, H.; Luquette, L.J.; Park, P.J. Detecting Somatic Mutations in Normal Cells. Trends Genet. 2018, 34, 545–557. [Google Scholar] [CrossRef]
- Gruber, A.J.; Gypas, F.; Riba, A.; Schmidt, R.; Zavolan, M. Terminal exon characterization with TECtool reveals an abundance of cell-specific isoforms. Nat. Methods 2018, 15, 832–836. [Google Scholar] [CrossRef]
- Kishore, S.; Luber, S.; Zavolan, M. Deciphering the role of RNA-binding proteins in the post-transcriptional control of gene expression. Brief. Funct. Genom. 2010, 9, 391–404. [Google Scholar] [CrossRef] [Green Version]
- Hausser, J.; Zavolan, M. Identification and consequences of miRNA-target interactions-beyond repression of gene expression. Nat. Rev. Genet. 2014, 15, 599–612. [Google Scholar] [CrossRef]
- La Manno, G.; Soldatov, R.; Zeisel, A.; Braun, E.; Hochgerner, H.; Petukhov, V.; Lidschreiber, K.; Kastriti, M.E.; Lönnerberg, P.; Furlan, A.; et al. RNA velocity of single cells. Nature 2018, 560, 494–498. [Google Scholar] [CrossRef] [Green Version]



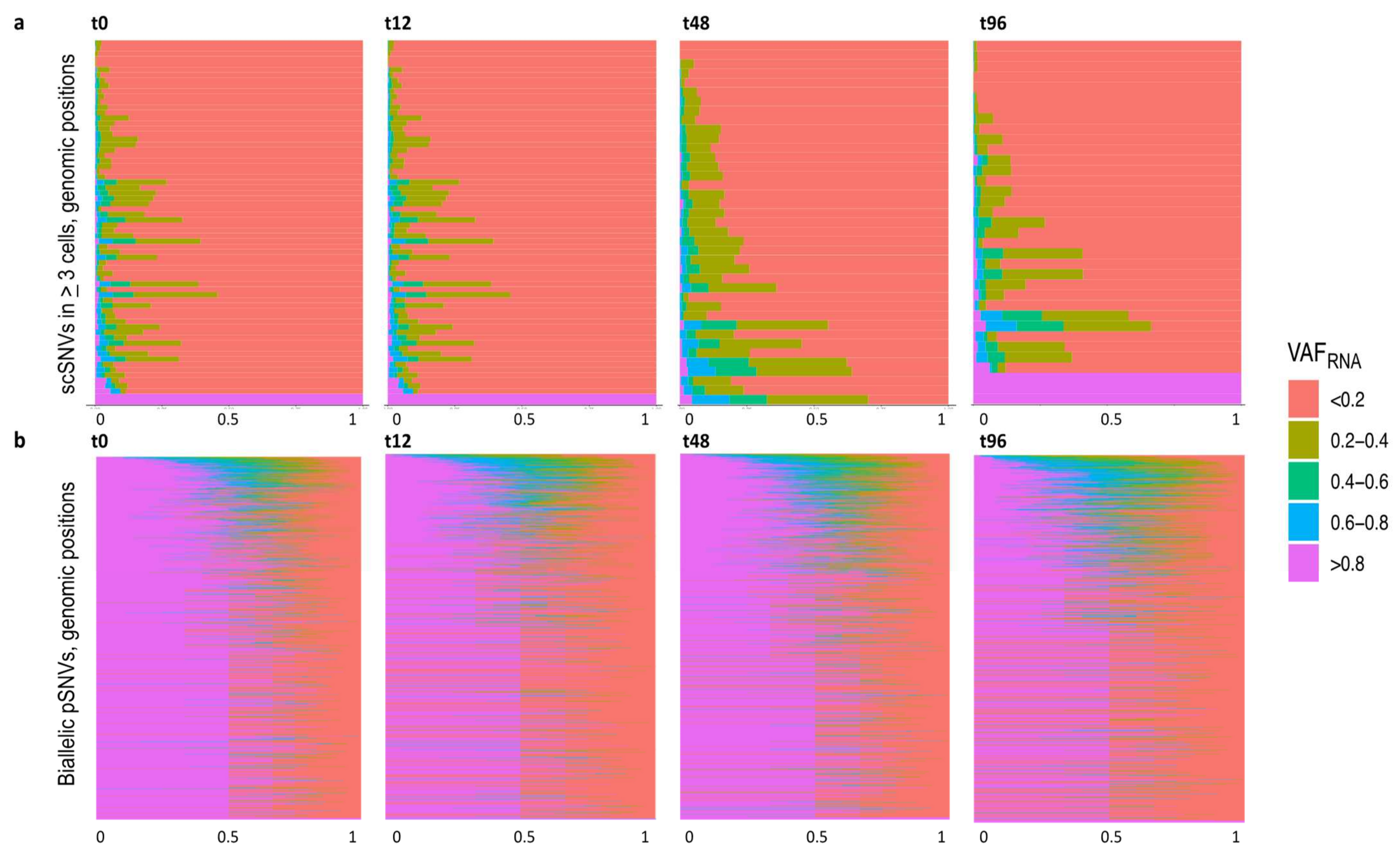
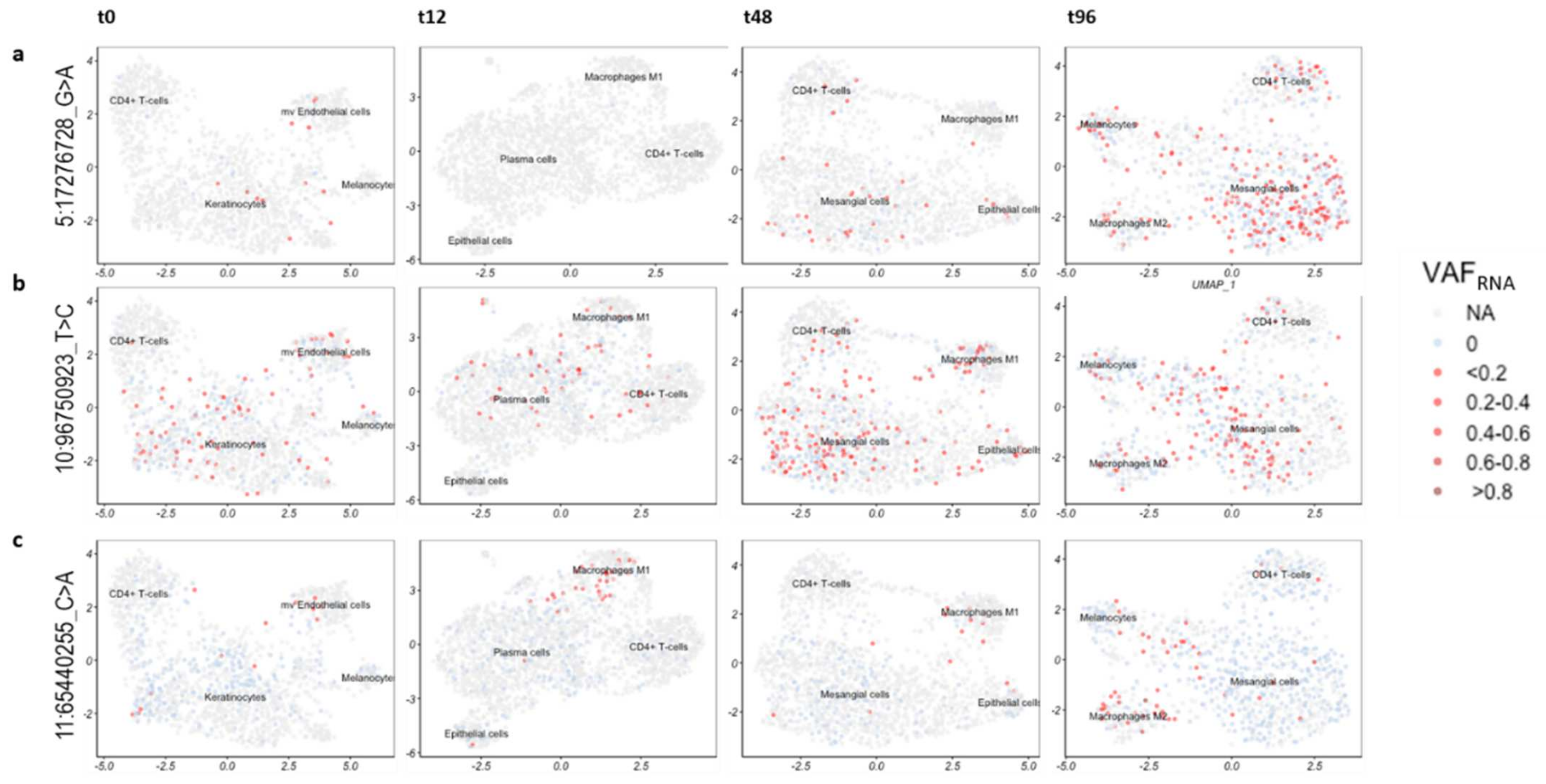
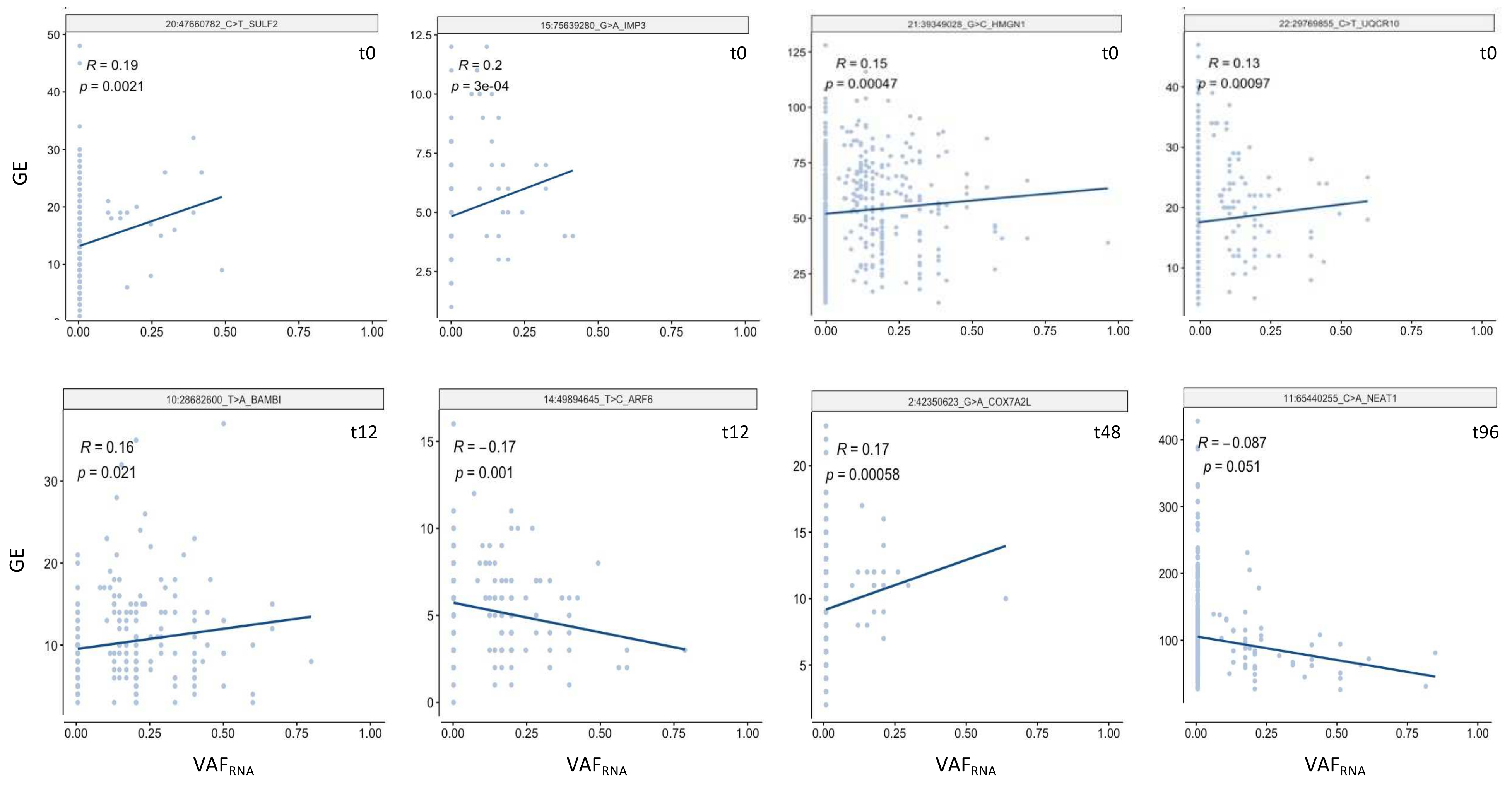
| Sample and Sequencing Approach | SRA id | N Cells | N SNVs GATK | N SNVs Strelka2 | Bulk/Pooled GATK and Strelka2 (Union) | Sc Alignments GATK and Strelka2 (Intersection) | N sceSNVs | N sceSNVs by Mutect2 |
|---|---|---|---|---|---|---|---|---|
| TES (POPv.3) | SRR5945460 | na | 395 | 390 | 409 | na | na | na |
| WGS | SRR5945478 | na | 25 | 322 | 322 | na | na | na |
| scRNA-seq t0 | SRR10018149 | 1749 | 256 | 2312 | 2800 * | 149 | 73 (49%) | 6 (8%) |
| scRNA-seq t12 | SRR10018150 | 2778 | 347 | 3373 | 3882 * | 101 | 61 (60%) | 7 (11%) |
| scRNA-seq t48 | SRR10018151 | 1891 | 218 | 2639 | 3132 * | 79 | 38 (48%) | 1 (3%) |
| scRNA-seq t96 | SRR10018152 | 1250 | 126 | 1481 | 1996 * | 86 | 45 (52%) | 2 (4%) |
| Sample | Pooled scRNA-seq Alignments (GATK and Strelka2 Union) | N sceSNVs (GATK and Strelka2 Intersection) | N sceSNVs by Mutect2 | Max Percent Cells with sceSNVs | N sceSNVs in 2 and More Cells |
|---|---|---|---|---|---|
| scRNA-seq t0 | 489,048 | 13,385 | 1310 (9.8%) | 90/1749 (5.2%) | 636 (4.8%) |
| scRNA-seq t12 | 524,598 | 9470 | 936 (9.9%) | 44/2778 (1.6%) | 472 (5%) |
| scRNA-seq t48 | 446,779 | 7131 | 560 (7.8%) | 33/1891 (1.8%) | 318 (4.5%) |
| scRNA-seq t96 | 335,839 | 10,794 | 856 (7.9%) | 30/1250 (2.4%) | 429 (4%) |
| Function | t0 | t12 | t48 | t96 | ||||
|---|---|---|---|---|---|---|---|---|
| sceSNVs | pSNVs | sceSNVs | pSNVs | sceSNVs | pSNVs | sceSNVs | pSNVs | |
| 3-prime-UTR | 8075 (60%) | 6062 (15%) | 5785 (61%) | 6340 (18%) | 4109 (57%) | 5618 (17%) | 6562 (61%) | 5272 (19%) |
| chi-square p-value | 10,883 p < 10−7 | 6981 p < 10−7 | 5092 p < 10−7 | 6403 p < 10−7 | ||||
| missense | 2010 (15%) | 1129 (3%) | 1562 (17%) | 1513 (4%) | 1323 (18%) | 1388 (4%) | 1744 (16%) | 915 (3%) |
| chi-square p-value | 2808 p < 10−7 | 1726 p < 10−7 | 1854 p < 10−7 | 2008 p < 10−7 | ||||
| intron | 1609 (12%) | 24,634 (60%) | 870 (9%) | 19,247 (55%) | 667 (9%) | 17,559 (54%) | 1045 (10%) | 14,351 (52%) |
| chi-square p-value | 9236 p < 10−7 | 6261 p < 10−7 | 4749 p < 10−7 | 5694 p < 10−7 | ||||
| synonymous | 897 (7%) | 1365 (3%) | 758 (8%) | 1844 (5%) | 580 (8%) | 1756 (5%) | 793 (7%) | 1213 (4%) |
| chi-square p-value | 287 p < 10−7 | 102 p < 10−7 | 76 p < 10−7 | 140 p < 10−7 | ||||
| intergenic | 276 (2%) | 6484 (16%) | 182 (2%) | 4680 (13%) | 161 (2%) | 4475 (14%) | 236 (2%) | 4941 (18%) |
| chi-square p-value | 1751 p < 10−7 | 997 p < 10−7 | 755 p < 10−7 | 1624 p < 10−7 | ||||
| non-coding exon | 250 (1.9%) | 414 (1%) | 142 (1.5%) | 357 (1%) | 102 (1.4%) | 331 (1%) | 200 (1.9%) | 421 (1.5%) |
| chi-square p-value | 60 p < 10−7 | 15 p = 0.00007 | 9 p = 0.002 | 6 p = 0.02 | ||||
| 5-prime-UTR | 152 (1.1%) | 816 (2%) | 78 (0.8%) | 1027 (2.9%) | 118 (1.6%) | 1098 (3.4%) | 80 (0.7%) | 622 (2.2%) |
| chi-square p-value | 41 p < 10−7 | 135 p < 10−7 | 58 p < 10−7 | 96 p < 10−7 | ||||
| splice | 16 (0.12%) | 105 (0.26%) | 15 (0.16%) | 120 (0.34%) | 9 (0.13%) | 103 (0.32%) | 18 (0.17%) | 78 (0.28%) |
| chi-square p-value | 8 p = 0.005 | 8 p = 0.006 | 7 p = 0.008 | 4 p = 0.04 | ||||
| stop | 100 (0.75%) | 6 (0.01%) | 78 (0.82%) | 6 (0.02%) | 62 (0.86%) | 2 (0.01%) | 116 (%1.07) | 5 (0.02%) |
| chi-square p-value | 275 p < 10−7 | 253 p < 10−7 | 264 p < 10−7 | 275 p < 10−7 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
N. M., P.; Liu, H.; Dillard, C.; Ibeawuchi, H.; Alsaeedy, T.; Chan, H.; Horvath, A.D. Improved SNV Discovery in Barcode-Stratified scRNA-seq Alignments. Genes 2021, 12, 1558. https://0-doi-org.brum.beds.ac.uk/10.3390/genes12101558
N. M. P, Liu H, Dillard C, Ibeawuchi H, Alsaeedy T, Chan H, Horvath AD. Improved SNV Discovery in Barcode-Stratified scRNA-seq Alignments. Genes. 2021; 12(10):1558. https://0-doi-org.brum.beds.ac.uk/10.3390/genes12101558
Chicago/Turabian StyleN. M., Prashant, Hongyu Liu, Christian Dillard, Helen Ibeawuchi, Turkey Alsaeedy, Hang Chan, and Anelia Dafinova Horvath. 2021. "Improved SNV Discovery in Barcode-Stratified scRNA-seq Alignments" Genes 12, no. 10: 1558. https://0-doi-org.brum.beds.ac.uk/10.3390/genes12101558






