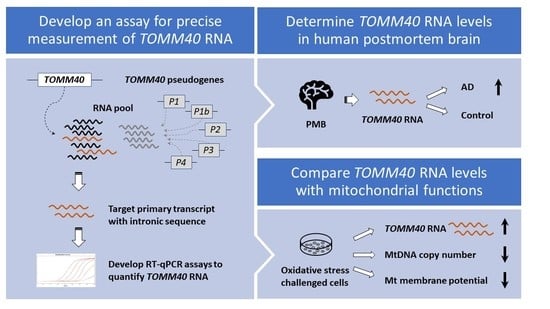
TOMM40 RNA Transcription in Alzheimer’s Disease Brain and Its Implication in Mitochondrial Dysfunction
Abstract
:1. Introduction
2. Materials and Methods
2.1. Human PMB and Cell Lines
2.2. DNA/RNA Extraction and Genotyping
2.3. Sequence Alignment and Phylogenetic Tree
2.4. Conventional End-Point PCR and Gel Electrophoresis
2.5. Reverse Transcriptase (RT) Reaction and Quantitative PCR (qPCR) Assay
2.6. Fraction Estimation of Pseudogene RNA and Surrogate RNA Using Digital PCR (dPCR)
2.7. Hydorogen Peroxide Treatment
2.8. MtDNA Copy Number Assay
2.9. Mitochondrial Membrane Potential (MMP) Assay
2.10. Statistical Analyses
3. Results
3.1. Presence of Pseudogene RNAs Obscures Accurate Measurement of TOMM40 mRNA
3.2. Development of TOMM40-Specific RT-PCR Assays
3.3. Effects of TOMM40 RNA Transcription Levels
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Takei, N.; Miyashita, A.; Tsukie, T.; Arai, H.; Asada, T.; Imagawa, M.; Shoji, M.; Higuchi, S.; Urakami, K.; Kimura, H.; et al. Genetic association study on in and around the APOE in late-onset Alzheimer disease in Japanese. Genomics 2009, 93, 441–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lambert, J.C.; Ibrahim-Verbaas, C.A.; Harold, D.; Naj, A.C.; Sims, R.; Bellenguez, C.; DeStafano, A.L.; Bis, J.C.; Beecham, G.W.; Grenier-Boley, B.; et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 2013, 45, 1452–1458. [Google Scholar] [CrossRef] [Green Version]
- Beecham, G.W.; Hamilton, K.; Naj, A.C.; Martin, E.R.; Huentelman, M.; Myers, A.J.; Corneveaux, J.J.; Hardy, J.; Vonsattel, J.P.; Younkin, S.G.; et al. Genome-wide association meta-analysis of neuropathologic features of Alzheimer’s disease and related dementias. PLoS Genet. 2014, 10, e1004606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yashin, A.I.; Fang, F.; Kovtun, M.; Wu, D.; Duan, M.; Arbeev, K.; Akushevich, I.; Kulminski, A.; Culminskaya, I.; Zhbannikov, I.; et al. Hidden heterogeneity in Alzheimer’s disease: Insights from genetic association studies and other analyses. Exp. Gerontol. 2018, 107, 148–160. [Google Scholar] [CrossRef] [PubMed]
- Sillen, A.; Forsell, C.; Lilius, L.; Axelman, K.; Bjork, B.F.; Onkamo, P.; Kere, J.; Winblad, B.; Graff, C. Genome scan on Swedish Alzheimer’s disease families. Mol. Psychiatry 2005, 11, 182–186. [Google Scholar] [CrossRef]
- Hendrie, H.C.; Murrell, J.; Baiyewu, O.; Lane, K.A.; Purnell, C.; Ogunniyi, A.; Unverzagt, F.W.; Hall, K.; Callahan, C.M.; Saykin, A.J.; et al. APOE epsilon4 and the risk for Alzheimer disease and cognitive decline in African Americans and Yoruba. Int. Psychogeriatr. 2014, 26, 977–985. [Google Scholar] [CrossRef] [Green Version]
- Rajabli, F.; Feliciano, B.E.; Celis, K.; Hamilton-Nelson, K.L.; Whitehead, P.L.; Adams, L.D.; Bussies, P.L.; Manrique, C.P.; Rodriguez, A.; Rodriguez, V.; et al. Ancestral origin of ApoE epsilon4 Alzheimer disease risk in Puerto Rican and African American populations. PLoS Genet. 2018, 14, e1007791. [Google Scholar] [CrossRef] [Green Version]
- Morris, J.C.; Schindler, S.E.; McCue, L.M.; Moulder, K.L.; Benzinger, T.L.S.; Cruchaga, C.; Fagan, A.M.; Grant, E.; Gordon, B.A.; Holtzman, D.M.; et al. Assessment of Racial Disparities in Biomarkers for Alzheimer Disease. JAMA Neurol. 2019. [Google Scholar] [CrossRef]
- Humphries, A.D.; Streimann, I.C.; Stojanovski, D.; Johnston, A.J.; Yano, M.; Hoogenraad, N.J.; Ryan, M.T. Dissection of the Mitochondrial Import and Assembly Pathway for Human Tom40. J. Biol. Chem. 2005, 280, 11535–11543. [Google Scholar] [CrossRef] [Green Version]
- Hoogenraad, N.J.; Ward, L.A.; Ryan, M.T. Import and assembly of proteins into mitochondria of mammalian cells. Biochim. Biophys. Acta 2002, 1592, 97–105. [Google Scholar] [CrossRef] [Green Version]
- Swerdlow, R.H.; Burns, J.M.; Khan, S.M. The Alzheimer’s disease mitochondrial cascade hypothesis: Progress and perspectives. Biochim. Biophys. Acta 2014, 1842, 1219–1231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwong, J.Q.; Beal, M.F.; Manfredi, G. The role of mitochondria in inherited neurodegenerative diseases. J. Neurochem. 2006, 97, 1659–1675. [Google Scholar] [CrossRef]
- Kapogiannis, D.; Mattson, M.P. Disrupted energy metabolism and neuronal circuit dysfunction in cognitive impairment and Alzheimer’s disease. Lancet Neurol. 2011, 10, 187–198. [Google Scholar] [CrossRef] [Green Version]
- Agrawal, I.; Jha, S. Mitochondrial Dysfunction and Alzheimer’s Disease: Role of Microglia. Front. Aging Neurosci. 2020, 12, 252. [Google Scholar] [CrossRef]
- Hirai, K.; Aliev, G.; Nunomura, A.; Fujioka, H.; Russell, R.L.; Atwood, C.S.; Johnson, A.B.; Kress, Y.; Vinters, H.V.; Tabaton, M.; et al. Mitochondrial abnormalities in Alzheimer’s disease. J. Neurosci. 2001, 21, 3017–3023. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Perry, G.; Moreira, P.I.; Aliev, G.; Cash, A.D.; Hirai, K.; Smith, M.A. Mitochondrial abnormalities and oxidative imbalance in Alzheimer disease. J. Alzheimers Dis. 2006, 9, 147–153. [Google Scholar] [CrossRef]
- Reddy, P.H.; Beal, M.F. Amyloid beta, mitochondrial dysfunction and synaptic damage: Implications for cognitive decline in aging and Alzheimer’s disease. Trends Mol. Med. 2008, 14, 45–53. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Su, B.; Zheng, L.; Perry, G.; Smith, M.A.; Zhu, X. The role of abnormal mitochondrial dynamics in the pathogenesis of Alzheimer’s disease. J. Neurochem. 2009, 109, 153–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Z.; Yang, Y.; Xiao, Z.; Zhao, Q.; Wu, W.; Liang, X.; Luo, J.; Cao, Y.; Shao, M.; Guo, Q.; et al. TOMM40 and APOE variants synergistically increase the risk of Alzheimer’s disease in a Chinese population. Aging Clin. Exp. Res. 2020. [Google Scholar] [CrossRef]
- Roses, A.D. An inherited variable poly-T repeat genotype in TOMM40 in Alzheimer disease. Arch. Neurol. 2010, 67, 536–541. [Google Scholar] [CrossRef]
- Lutz, M.W.; Crenshaw, D.G.; Saunders, A.M.; Roses, A.D. Genetic variation at a single locus and age of onset for Alzheimer’s disease. Alzheimers Dement. 2010, 6, 125–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lutz, M.W.; Sundseth, S.S.; Burns, D.K.; Saunders, A.M.; Hayden, K.M.; Burke, J.R.; Welsh-Bohmer, K.A.; Roses, A.D. A Genetics-based Biomarker Risk Algorithm for Predicting Risk of Alzheimer’s Disease. Alzheimers Dement. 2016, 2, 30–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crenshaw, D.G.; Gottschalk, W.K.; Lutz, M.W.; Grossman, I.; Saunders, A.M.; Burke, J.R.; Welsh-Bohmer, K.A.; Brannan, S.K.; Burns, D.K.; Roses, A.D. Using genetics to enable studies on the prevention of Alzheimer’s disease. Clin. Pharm. 2013, 93, 177–185. [Google Scholar] [CrossRef] [Green Version]
- Linnertz, C.; Anderson, L.; Gottschalk, W.; Crenshaw, D.; Lutz, M.W.; Allen, J.; Saith, S.; Mihovilovic, M.; Burke, J.R.; Welsh-Bohmer, K.A.; et al. The cis-regulatory effect of an Alzheimer’s disease-associated poly-T locus on expression of TOMM40 and apolipoprotein E genes. Alzheimers Dement. 2014, 10, 541–551. [Google Scholar] [CrossRef] [Green Version]
- Yu, C.E.; Seltman, H.; Peskind, E.R.; Galloway, N.; Zhou, P.X.; Rosenthal, E.; Wijsman, E.M.; Tsuang, D.W.; Devlin, B.; Schellenberg, G.D. Comprehensive analysis of APOE and selected proximate markers for late-onset Alzheimer’s disease: Patterns of linkage disequilibrium and disease/marker association. Genomics 2007, 89, 655–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blue, E.E.; Cheng, A.; Chen, S.; Yu, C.E.; Alzheimer’s Disease Genetics, C. Association of Uncommon, Noncoding Variants in the APOE Region With Risk of Alzheimer Disease in Adults of European Ancestry. JAMA Netw. Open 2020, 3, e2017666. [Google Scholar] [CrossRef]
- Bekris, L.M.; Millard, S.P.; Galloway, N.M.; Vuletic, S.; Albers, J.J.; Li, G.; Galasko, D.R.; DeCarli, C.; Farlow, M.R.; Clark, C.M.; et al. Multiple SNPs within and surrounding the apolipoprotein E gene influence cerebrospinal fluid apolipoprotein E protein levels. J. Alzheimers Dis. 2008, 13, 255–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bekris, L.M.; Galloway, N.M.; Montine, T.J.; Schellenberg, G.D.; Yu, C.E. APOE mRNA and protein expression in postmortem brain are modulated by an extended haplotype structure. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2009. [Google Scholar] [CrossRef] [Green Version]
- Bekris, L.M.; Lutz, F.; Yu, C.-E. Functional analysis of APOE locus genetic variation implicates regional enhancers in the regulation of both TOMM40 and APOE. J. Hum. Genet. 2012, 57, 18–25. [Google Scholar] [CrossRef] [Green Version]
- Hansson Petersen, C.A.; Alikhani, N.; Behbahani, H.; Wiehager, B.; Pavlov, P.F.; Alafuzoff, I.; Leinonen, V.; Ito, A.; Winblad, B.; Glaser, E.; et al. The amyloid beta-peptide is imported into mitochondria via the TOM import machinery and localized to mitochondrial cristae. Proc. Natl. Acad. Sci. USA 2008, 105, 13145–13150. [Google Scholar] [CrossRef] [Green Version]
- Anandatheerthavarada, H.K.; Devi, L. Mitochondrial translocation of amyloid precursor protein and its cleaved products: Relevance to mitochondrial dysfunction in Alzheimer’s disease. Rev. Neurosci. 2007, 18, 343–354. [Google Scholar] [CrossRef] [PubMed]
- Cenini, G.; Rub, C.; Bruderek, M.; Voos, W. Amyloid beta-peptides interfere with mitochondrial preprotein import competence by a coaggregation process. Mol. Biol. Cell 2016, 27, 3257–3272. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, P.M.; von Einem, B.; Walther, P.; Calzia, E.; von Arnim, C.A. Metabolic Characterization of Intact Cells Reveals Intracellular Amyloid Beta but Not Its Precursor Protein to Reduce Mitochondrial Respiration. PLoS ONE 2016, 11, e0168157. [Google Scholar] [CrossRef]
- Devi, L.; Prabhu, B.M.; Galati, D.F.; Avadhani, N.G.; Anandatheerthavarada, H.K. Accumulation of amyloid precursor protein in the mitochondrial import channels of human Alzheimer’s disease brain is associated with mitochondrial dysfunction. J. Neurosci. 2006, 26, 9057–9068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mise, A.; Yoshino, Y.; Yamazaki, K.; Ozaki, Y.; Sao, T.; Yoshida, T.; Mori, T.; Mori, Y.; Ochi, S.; Iga, J.I.; et al. TOMM40 and APOE Gene Expression and Cognitive Decline in Japanese Alzheimer’s Disease Subjects. J. Alzheimers Dis. 2017, 60, 1107–1117. [Google Scholar] [CrossRef]
- Chong, M.S.; Goh, L.K.; Lim, W.S.; Chan, M.; Tay, L.; Chen, G.; Feng, L.; Ng, T.P.; Tan, C.H.; Lee, T.S. Gene expression profiling of peripheral blood leukocytes shows consistent longitudinal downregulation of TOMM40 and upregulation of KIR2DL5A, PLOD1, and SLC2A8 among fast progressors in early Alzheimer’s disease. J. Alzheimers Dis. 2013, 34, 399–405. [Google Scholar] [CrossRef]
- Lee, T.S.; Goh, L.; Chong, M.S.; Chua, S.M.; Chen, G.B.; Feng, L.; Lim, W.S.; Chan, M.; Ng, T.P.; Krishnan, K.R. Downregulation of TOMM40 expression in the blood of Alzheimer disease subjects compared with matched controls. J. Psychiatr Res. 2012, 46, 828–830. [Google Scholar] [CrossRef]
- Goh, L.K.; Lim, W.S.; Teo, S.; Vijayaraghavan, A.; Chan, M.; Tay, L.; Ng, T.P.; Tan, C.H.; Lee, T.S.; Chong, M.S. TOMM40 alterations in Alzheimer’s disease over a 2-year follow-up period. J. Alzheimers Dis. 2015, 44, 57–61. [Google Scholar] [CrossRef]
- Manczak, M.; Calkins, M.J.; Reddy, P.H. Impaired mitochondrial dynamics and abnormal interaction of amyloid beta with mitochondrial protein Drp1 in neurons from patients with Alzheimer’s disease: Implications for neuronal damage. Hum. Mol. Genet. 2011, 20, 2495–2509. [Google Scholar] [CrossRef]
- Lee, E.G.; Tulloch, J.; Chen, S.; Leong, L.; Saxton, A.D.; Kraemer, B.; Darvas, M.; Keene, C.D.; Shutes-David, A.; Todd, K.; et al. Redefining transcriptional regulation of the APOE gene and its association with Alzheimer’s disease. PLoS ONE 2020, 15, e0227667. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Witek, J.; Mohiuddin, S.S. Biochemistry, Pseudogenes. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Cheng, A.; Hou, Y.; Mattson, M.P. Mitochondria and neuroplasticity. ASN Neuro 2010, 2, e00045. [Google Scholar] [CrossRef]
- Markham, A.; Bains, R.; Franklin, P.; Spedding, M. Changes in mitochondrial function are pivotal in neurodegenerative and psychiatric disorders: How important is BDNF? Br. J. Pharm. 2014, 171, 2206–2229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roses, A.; Sundseth, S.; Saunders, A.; Gottschalk, W.; Burns, D.; Lutz, M. Understanding the genetics of APOE and TOMM40 and role of mitochondrial structure and function in clinical pharmacology of Alzheimer’s disease. Alzheimers Dement. 2016, 12, 687–694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, E.R.; Lai, E.H.; Gilbert, J.R.; Rogala, A.R.; Afshari, A.J.; Riley, J.; Finch, K.L.; Stevens, J.F.; Livak, K.J.; Slotterbeck, B.D.; et al. SNPing away at complex diseases: Analysis of single-nucleotide polymorphisms around APOE in Alzheimer disease. Am. J. Hum. Genet. 2000, 67, 383–394. [Google Scholar] [CrossRef] [Green Version]
- Martin, E.R.; Gilbert, J.R.; Lai, E.H.; Riley, J.; Rogala, A.R.; Slotterbeck, B.D.; Sipe, C.A.; Grubber, J.M.; Warren, L.L.; Conneally, P.M.; et al. Analysis of association at single nucleotide polymorphisms in the APOE region. Genomics 2000, 63, 7–12. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Pierce, B.L. Genetic susceptibility to accelerated cognitive decline in the US Health and Retirement Study. Neurobiol. Aging 2014, 35, 1512.e11–1512.e18. [Google Scholar] [CrossRef]
- Davies, G.; Armstrong, N.; Bis, J.C.; Bressler, J.; Chouraki, V.; Giddaluru, S.; Hofer, E.; Ibrahim-Verbaas, C.A.; Kirin, M.; Lahti, J.; et al. Genetic contributions to variation in general cognitive function: A meta-analysis of genome-wide association studies in the CHARGE consortium (N = 53949). Mol. Psychiatry 2015, 20, 183–192. [Google Scholar] [CrossRef] [Green Version]
- Fortney, K.; Dobriban, E.; Garagnani, P.; Pirazzini, C.; Monti, D.; Mari, D.; Atzmon, G.; Barzilai, N.; Franceschi, C.; Owen, A.B.; et al. Genome-Wide Scan Informed by Age-Related Disease Identifies Loci for Exceptional Human Longevity. PLoS Genet. 2015, 11, e1005728. [Google Scholar] [CrossRef]
- Liu, X.; Song, Z.; Li, Y.; Yao, Y.; Fang, M.; Bai, C.; An, P.; Chen, H.; Chen, Z.; Tang, B.; et al. Integrated genetic analyses revealed novel human longevity loci and reduced risks of multiple diseases in a cohort study of 15,651 Chinese individuals. Aging Cell 2021, 20, e13323. [Google Scholar] [CrossRef] [PubMed]
- Deelen, J.; Beekman, M.; Uh, H.W.; Helmer, Q.; Kuningas, M.; Christiansen, L.; Kremer, D.; van der Breggen, R.; Suchiman, H.E.; Lakenberg, N.; et al. Genome-wide association study identifies a single major locus contributing to survival into old age; the APOE locus revisited. Aging Cell 2011, 10, 686–698. [Google Scholar] [CrossRef]
- Guo, Y.; Lanktree, M.B.; Taylor, K.C.; Hakonarson, H.; Lange, L.A.; Keating, B.J.; Consortium, I.K.S. Gene-centric meta-analyses of 108 912 individuals confirm known body mass index loci and reveal three novel signals. Hum. Mol. Genet. 2013, 22, 184–201. [Google Scholar] [CrossRef] [Green Version]
- Middelberg, R.P.; Ferreira, M.A.; Henders, A.K.; Heath, A.C.; Madden, P.A.; Montgomery, G.W.; Martin, N.G.; Whitfield, J.B. Genetic variants in LPL, OASL and TOMM40/APOE-C1-C2-C4 genes are associated with multiple cardiovascular-related traits. BMC Med. Genet. 2011, 12, 123. [Google Scholar] [CrossRef] [Green Version]
- Sandhu, M.S.; Waterworth, D.M.; Debenham, S.L.; Wheeler, E.; Papadakis, K.; Zhao, J.H.; Song, K.; Yuan, X.; Johnson, T.; Ashford, S.; et al. LDL-cholesterol concentrations: A genome-wide association study. Lancet 2008, 371, 483–491. [Google Scholar] [CrossRef] [Green Version]
- Denny, J.C.; Bastarache, L.; Ritchie, M.D.; Carroll, R.J.; Zink, R.; Mosley, J.D.; Field, J.R.; Pulley, J.M.; Ramirez, A.H.; Bowton, E.; et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat. Biotechnol. 2013, 31, 1102–1110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roses, A.D.; Lutz, M.W.; Crenshaw, D.G.; Grossman, I.; Saunders, A.M.; Gottschalk, W.K. TOMM40 and APOE: Requirements for replication studies of association with age of disease onset and enrichment of a clinical trial. Alzheimers Dement. 2013, 9, 132–136. [Google Scholar] [CrossRef] [Green Version]
- Zheng, H.; Xie, W. The role of 3D genome organization in development and cell differentiation. Nat. Rev. Mol. Cell Biol. 2019, 20, 535–550. [Google Scholar] [CrossRef] [PubMed]
- Leek, B.F. Abdominal and pelvic visceral receptors. Br. Med. Bull. 1977, 33, 163–168. [Google Scholar] [CrossRef]
- Dowling, P.; Meleady, P.; Dowd, A.; Henry, M.; Glynn, S.; Clynes, M. Proteomic analysis of isolated membrane fractions from superinvasive cancer cells. Biochim. Biophys. Acta 2007, 1774, 93–101. [Google Scholar] [CrossRef]
- Heinemeyer, T.; Stemmet, M.; Bardien, S.; Neethling, A. Underappreciated Roles of the Translocase of the Outer and Inner Mitochondrial Membrane Protein Complexes in Human Disease. DNA Cell Biol. 2019, 38, 23–40. [Google Scholar] [CrossRef] [PubMed]
- Bender, A.; Desplats, P.; Spencer, B.; Rockenstein, E.; Adame, A.; Elstner, M.; Laub, C.; Mueller, S.; Koob, A.O.; Mante, M.; et al. TOM40 mediates mitochondrial dysfunction induced by alpha-synuclein accumulation in Parkinson′s disease. PLoS ONE 2013, 8, e62277. [Google Scholar] [CrossRef] [Green Version]
- Valente, E.M.; Abou-Sleiman, P.M.; Caputo, V.; Muqit, M.M.; Harvey, K.; Gispert, S.; Ali, Z.; Del Turco, D.; Bentivoglio, A.R.; Healy, D.G.; et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science 2004, 304, 1158–1160. [Google Scholar] [CrossRef] [Green Version]
- Pakpian, N.; Phopin, K.; Kitidee, K.; Govitrapong, P.; Wongchitrat, P. Alterations in Mitochondrial Dynamic-related Genes in the Peripheral Blood of Alzheimer’s Disease Patients. Curr. Alzheimer Res. 2020, 17, 616–625. [Google Scholar] [CrossRef]
- Carrocci, T.J.; Neugebauer, K.M. Pre-mRNA Splicing in the Nuclear Landscape. Cold Spring Harb. Symp. Quant. Biol. 2019, 84, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Agirre, E.; Oldfield, A.J.; Bellora, N.; Segelle, A.; Luco, R.F. Splicing-associated chromatin signatures: A combinatorial and position-dependent role for histone marks in splicing definition. Nat. Commun. 2021, 12, 682. [Google Scholar] [CrossRef] [PubMed]
- Aslanzadeh, V.; Huang, Y.; Sanguinetti, G.; Beggs, J.D. Transcription rate strongly affects splicing fidelity and cotranscriptionality in budding yeast. Genome Res. 2018, 28, 203–213. [Google Scholar] [CrossRef] [Green Version]
- Abebrese, E.L.; Ali, S.H.; Arnold, Z.R.; Andrews, V.M.; Armstrong, K.; Burns, L.; Crowder, H.R.; Day, R.T., Jr.; Hsu, D.G.; Jarrell, K.; et al. Identification of human short introns. PLoS ONE 2017, 12, e0175393. [Google Scholar] [CrossRef]
- Kann, O.; Kovacs, R. Mitochondria and neuronal activity. Am. J. Physiol. Cell Physiol. 2007, 292, C641–C657. [Google Scholar] [CrossRef]
- Chan, D.C. Mitochondria: Dynamic organelles in disease, aging, and development. Cell 2006, 125, 1241–1252. [Google Scholar] [CrossRef] [Green Version]
- Castellani, C.A.; Longchamps, R.J.; Sun, J.; Guallar, E.; Arking, D.E. Thinking outside the nucleus: Mitochondrial DNA copy number in health and disease. Mitochondrion 2020, 53, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.Y.; Castellani, C.A.; Longchamps, R.J.; Pillalamarri, V.K.; O’Rourke, B.; Guallar, E.; Arking, D.E. Blood-derived mitochondrial DNA copy number is associated with gene expression across multiple tissues and is predictive for incident neurodegenerative disease. Genome Res. 2021, 31, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Longchamps, R.J.; Castellani, C.A.; Yang, S.Y.; Newcomb, C.E.; Sumpter, J.A.; Lane, J.; Grove, M.L.; Guallar, E.; Pankratz, N.; Taylor, K.D.; et al. Evaluation of mitochondrial DNA copy number estimation techniques. PLoS ONE 2020, 15, e0228166. [Google Scholar] [CrossRef] [Green Version]
- Filograna, R.; Mennuni, M.; Alsina, D.; Larsson, N.G. Mitochondrial DNA copy number in human disease: The more the better? FEBS Lett. 2020. [Google Scholar] [CrossRef]
- Coskun, P.E.; Beal, M.F.; Wallace, D.C. Alzheimer’s brains harbor somatic mtDNA control-region mutations that suppress mitochondrial transcription and replication. Proc. Natl. Acad. Sci. USA 2004, 101, 10726–10731. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Santiago, B.; Casademont, J.; Nunes, V. Is mitochondrial DNA depletion involved in Alzheimer’s disease? Eur. J. Hum. Genet. 2001, 9, 279–285. [Google Scholar] [CrossRef]
- Zamzami, N.; Marchetti, P.; Castedo, M.; Decaudin, D.; Macho, A.; Hirsch, T.; Susin, S.A.; Petit, P.X.; Mignotte, B.; Kroemer, G. Sequential reduction of mitochondrial transmembrane potential and generation of reactive oxygen species in early programmed cell death. J. Exp. Med. 1995, 182, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Yaniv, Y.; Juhaszova, M.; Nuss, H.B.; Wang, S.; Zorov, D.B.; Lakatta, E.G.; Sollott, S.J. Matching ATP supply and demand in mammalian heart: In vivo, in vitro, and in silico perspectives. Ann. N. Y. Acad. Sci 2010, 1188, 133–142. [Google Scholar] [CrossRef] [Green Version]
- Zorov, D.B.; Juhaszova, M.; Sollott, S.J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 2014, 94, 909–950. [Google Scholar] [CrossRef] [Green Version]
- Izyumov, D.S.; Avetisyan, A.V.; Pletjushkina, O.Y.; Sakharov, D.V.; Wirtz, K.W.; Chernyak, B.V.; Skulachev, V.P. “Wages of fear”: Transient threefold decrease in intracellular ATP level imposes apoptosis. Biochim. Biophys. Acta 2004, 1658, 141–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Subjects | AD | Control |
|---|---|---|
| Sample number—n | 47 | 20 |
| Gender—n female (% female) | 27 (57.4) | 11 (55.0) |
| APOE ε4+—n (%) | 29 (61.7) | 3 (15.0) |
| Age at death—mean (SD) | 87.9 (5.9) | 88.3 (8.5) |
| Age at onset—mean (SD) | 79.0 (8.0) | N/A |
| Disease duration—mean years (SD) | 9.0 (4.4) | N/A |
| Postmortem interval—mean hours (SD) | 5.0 (2.0) | 4.9 (2.3) |
| CERAD Score | ||
| Absent | 0 | 7 |
| Sparse | 0 | 7 |
| Moderate | 11 | 4 |
| Frequent | 36 | 2 |
| Braak Stage | ||
| I | 0 | 6 |
| II | 0 | 11 |
| III | 0 | 3 |
| IV | 0 | 0 |
| V | 15 | 0 |
| VI | 32 | 0 |
| Gene/Pseudogene | Genomic Sequence (hg38) | RNA Sequence (RefSeq) | |||
|---|---|---|---|---|---|
| Coordinate | Span (bp) | Accession # | Size (nt) | BLASTn ** | |
| TOMM40 | chr19: 44891220-44903689 | 12,470 | NM_001128916 | 1676 | - |
| TOMM40P1 | chr14: 19266948-19268660 | 1713 | NG_022836 | 1713 * | 95.70% |
| TOMM40P1b | chr14: 19131227-19133057 | 1831 | N/A | 1831 * | 95.55% |
| TOMM40P2 | chr22: 15853581-15855410 | 1830 | NG_022885 | 1830 * | 95.88% |
| TOMM40P3 | chr5: 3501872-33503327 | 1456 | NG_021878 | 1456 * | 87.26% |
| TOMM40P4 | chr2: 31723017-131724478 | 1462 | NG_023610 | 1462 * | 95.41% |
| TOMM40L | chr1: 161226060-161230746 | 4687 | NM_032174 | 2790 | 70.17% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, E.-G.; Chen, S.; Leong, L.; Tulloch, J.; Yu, C.-E. TOMM40 RNA Transcription in Alzheimer’s Disease Brain and Its Implication in Mitochondrial Dysfunction. Genes 2021, 12, 871. https://0-doi-org.brum.beds.ac.uk/10.3390/genes12060871
Lee E-G, Chen S, Leong L, Tulloch J, Yu C-E. TOMM40 RNA Transcription in Alzheimer’s Disease Brain and Its Implication in Mitochondrial Dysfunction. Genes. 2021; 12(6):871. https://0-doi-org.brum.beds.ac.uk/10.3390/genes12060871
Chicago/Turabian StyleLee, Eun-Gyung, Sunny Chen, Lesley Leong, Jessica Tulloch, and Chang-En Yu. 2021. "TOMM40 RNA Transcription in Alzheimer’s Disease Brain and Its Implication in Mitochondrial Dysfunction" Genes 12, no. 6: 871. https://0-doi-org.brum.beds.ac.uk/10.3390/genes12060871