Silencing of HMGA2 by siRNA Loaded Methotrexate Functionalized Polyamidoamine Dendrimer for Human Breast Cancer Cell Therapy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis of G4/MTX Nanosystem
2.3. Preparation of G4/MTX-siRNA Nanocomplex
2.4. Characterization f G4/MTX Nanosystem and G4/MTX-siRNA Nanocomplex
2.5. Gel Retardation Assay Electrophoresis
2.6. MTX and siRNA Release from the G4/MTX-siRNA Nanocomplex
2.7. Cell Culture and In Vitro Transfection Experiment
2.8. Cellular Uptake Study
2.9. Gene Expression
2.10. Western Blot Analysis
2.11. Cell Viability Assay
2.12. Apoptosis Assay
2.12.1. Annexin V/Propidium Iodide Staining
2.12.2. DAPI Staining
2.13. Statistical Analysis
3. Results
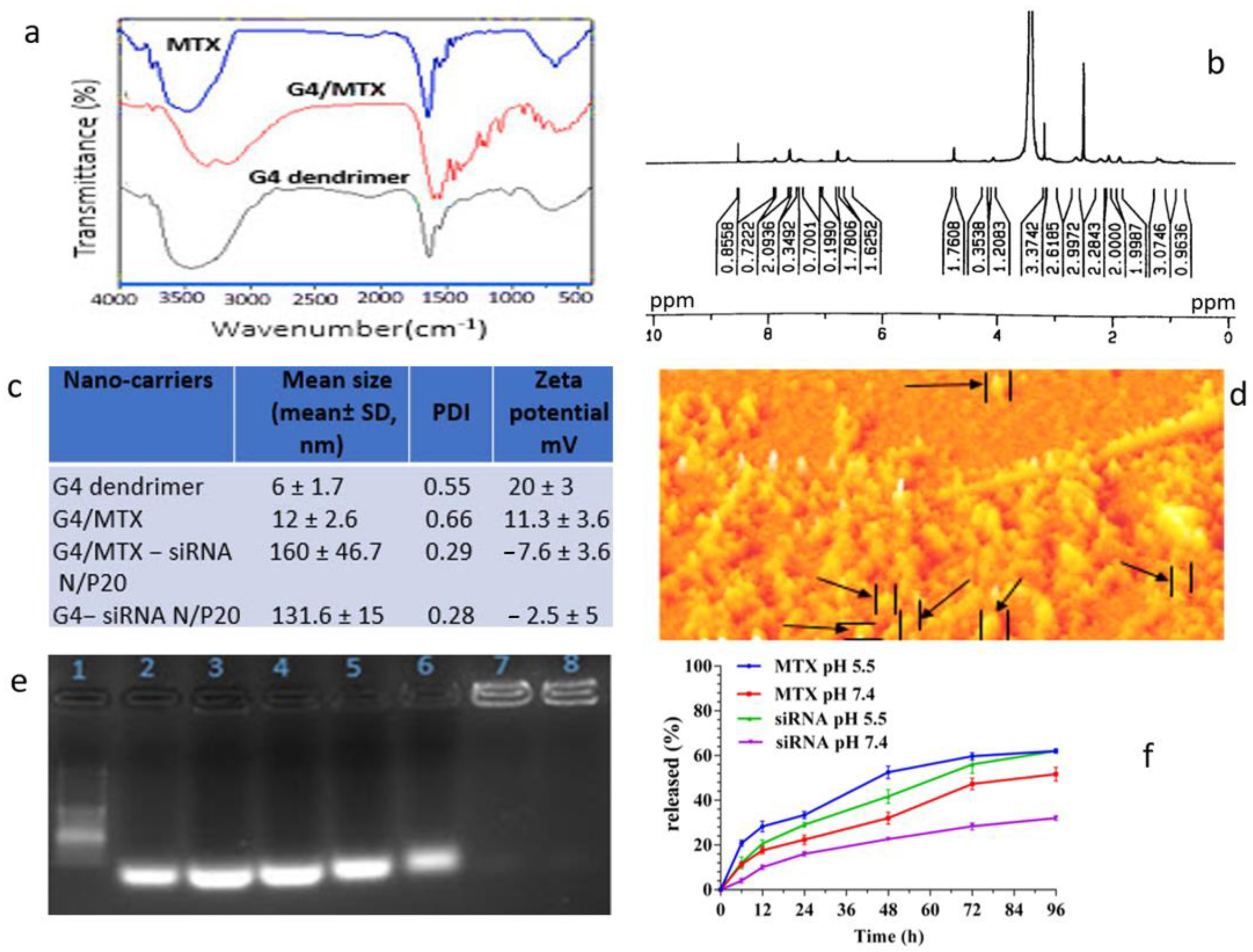
3.1. Confirmation of G4/MTX Nanosystem Synthesis
3.2. Characterization of G4/MTX Nanosystem and G4/MTX-siRNA Nanocomplex
3.3. HMGA2 Specific siRNA Electrostatically Loaded on the G4/MTX Nanosystem
3.4. MTX and HMGA2 siRNA Release Profile
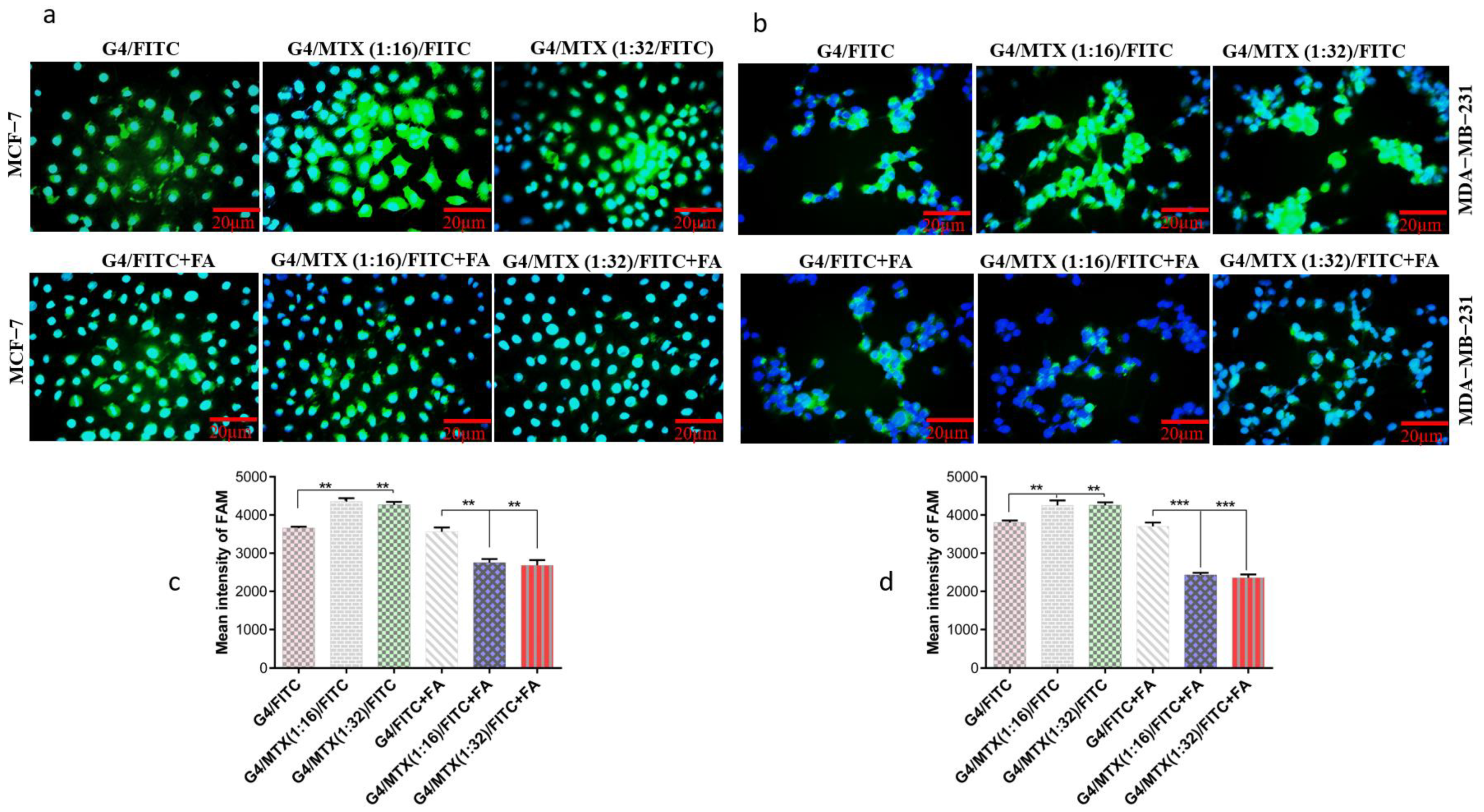
3.5. G4/MTX Nanosystem Efficiently Delivered HMGA2 siRNA into the FR Expressing Cancer Cells
3.6. HMGA2 Gene and Protein Expression Profile
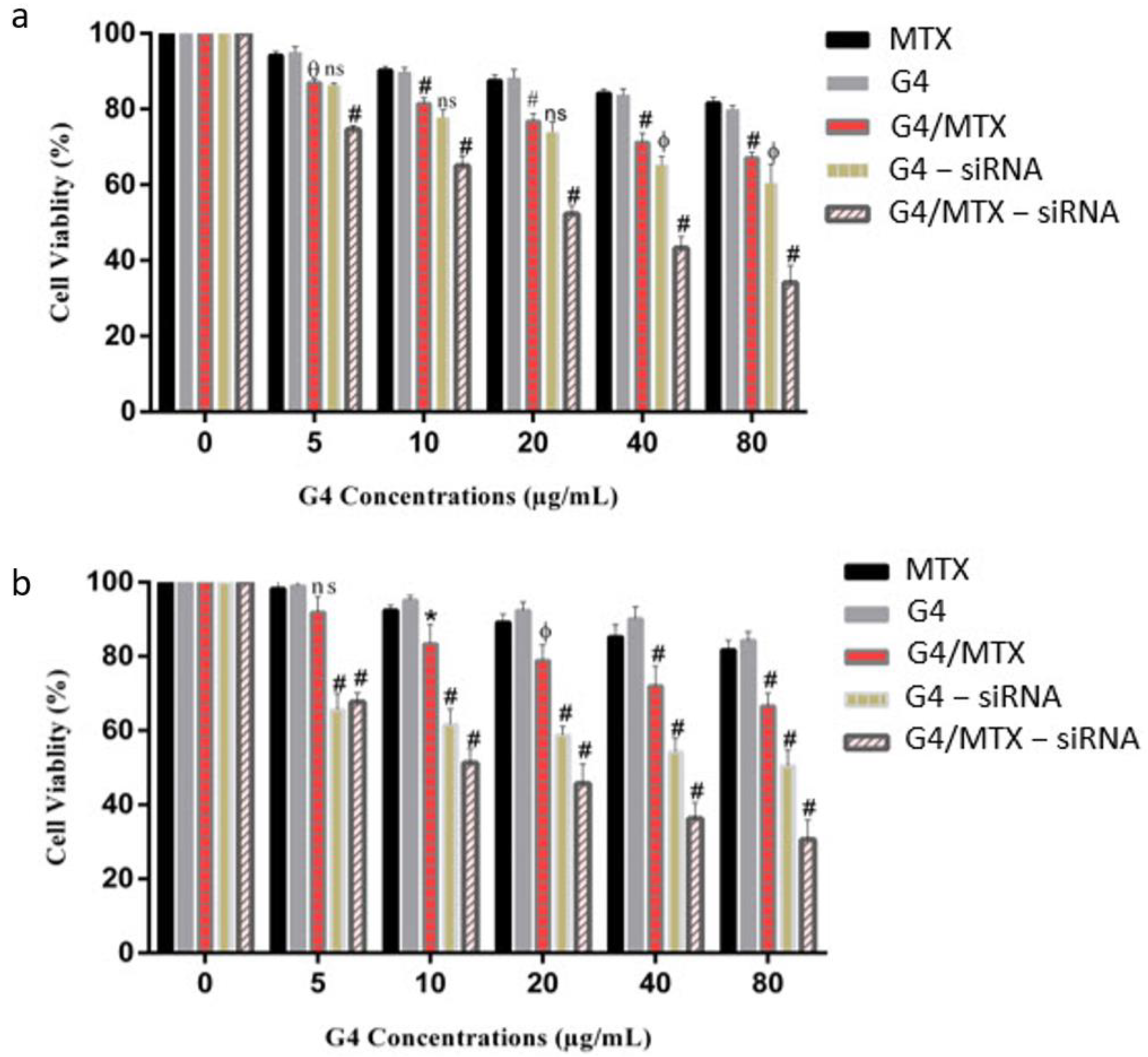
3.7. In Vitro Cytotoxicity Study
3.8. Suppression of HMGA2 Enhanced Cell Apoptosis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gaballu, F.A.; Dehghan, G.; Ghaffari, M.; Yekta, R.; Abbaspour-Ravasjani, S.; Baradaran, B.; Dolatabadi, J.E.N.; Hamblin, M.R. PAMAM dendrimers as efficient drug and gene delivery nanosystems for cancer therapy. Appl. Mater. Today 2018, 12, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Senapati, S.; Mahanta, A.K.; Kumar, S.; Maiti, P. Controlled drug delivery vehicles for cancer treatment and their performance. Signal Transduct. Target. Ther. 2018, 3, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Nami, S.; Aghebati-Maleki, A.; Aghebati-Maleki, L. Current applications and prospects of nanoparticles for antifungal drug delivery. EXCLI J. 2021, 20, 562–584. [Google Scholar]
- Chivere, V.T.; Kondiah, P.P.D.; Choonara, Y.E.; Pillay, V. Nanotechnology-Based Biopolymeric Oral Delivery Platforms for Advanced Cancer Treatment. Cancers 2020, 12, 522. [Google Scholar] [CrossRef] [Green Version]
- Zeinali, M.; Abbaspour-Ravasjani, S.; Ghorbani, M.; Babazadeh, A.; Soltanfam, T.; Santos, A.C.; Hamishehkar, H.; Hamblin, M.R. Nanovehicles for co-delivery of anticancer agents. Drug Discov. Today 2020, 25, 1416–1430. [Google Scholar] [CrossRef]
- Patra, J.K.; Das, G.; Fraceto, L.F.; Campos, E.V.R.; del Pilar Rodriguez-Torres, M.; Acosta-Torres, L.S.; Diaz-Torres, L.A.; Grillo, R.; Swamy, M.K.; Sharma, S.; et al. Nano based drug delivery systems: Recent developments and future prospects. J. Nanobiotechnol. 2018, 16, 71. [Google Scholar] [CrossRef] [Green Version]
- Truebenbach, I.; Gorges, J.; Kuhn, J.; Kern, S.; Baratti, E.; Kazmaier, U.; Wagner, E.; Lächelt, U. Sequence-Defined Oligoamide Drug Conjugates of Pretubulysin and Methotrexate for Folate Receptor Targeted Cancer Therapy. Macromol. Biosci. 2017, 17, 1600520. [Google Scholar] [CrossRef] [Green Version]
- Álvarez-González, B.; Rozalen, M.; Fernández-Perales, M.; Álvarez, M.A.; Sánchez-Polo, M. Methotrexate Gold Nanocarriers: Loading and Release Study: Its Activity in Colon and Lung Cancer Cells. Molecules 2020, 25, 6049. [Google Scholar] [CrossRef]
- Behzadi, S.; Serpooshan, V.; Tao, W.; Hamaly, M.A.; Alkawareek, M.Y.; Dreaden, E.C.; Brown, D.; Alkilany, A.M.; Farokhzad, O.C.; Mahmoudi, M. Cellular uptake of nanoparticles: Journey inside the cell. Chem. Soc. Rev. 2017, 46, 4218–4244. [Google Scholar] [CrossRef]
- Steinborn, B.; Truebenbach, I.; Morys, S.; Lächelt, U.; Wagner, E.; Zhang, W. Epidermal growth factor receptor targeted methotrexate and small interfering RNA co-delivery. J. Gene Med. 2018, 20, e3041. [Google Scholar] [CrossRef] [PubMed]
- Hao, F.; Lee, R.J.; Yang, C.; Zhong, L.; Sun, Y.; Dong, S.; Cheng, Z.; Teng, L.; Meng, Q.; Lu, J.; et al. Targeted Co-Delivery of siRNA and Methotrexate for Tumor Therapy via Mixed Micelles. Pharmaceutics 2019, 11, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dorsett, Y.; Tuschl, T. siRNAs: Applications in functional genomics and potential as therapeutics. Nat. Rev. Drug Discov. 2004, 3, 318–329. [Google Scholar] [CrossRef]
- Wittrup, A.; Lieberman, J. Knocking down disease: A progress report on siRNA therapeutics. Nat. Rev. Genet. 2015, 16, 543–552. [Google Scholar] [CrossRef] [PubMed]
- Nam, J.-P.; Nam, K.; Jung, S.; Nah, J.-W.; Kim, S.W. Evaluation of dendrimer type bio-reducible polymer as a siRNA delivery carrier for cancer therapy. J. Control. Release 2015, 209, 179–185. [Google Scholar] [CrossRef] [Green Version]
- Wilson, R.C.; Doudna, J.A. Molecular Mechanisms of RNA Interference. Annu. Rev. Biophys. 2013, 42, 217–239. [Google Scholar] [CrossRef] [Green Version]
- Mansoori, B.; Mohammadi, A.; Ditzel, H.J.; Duijf, P.H.; Khaze, V.; Gjerstorff, M.F.; Baradaran, B. HMGA2 as a Critical Regulator in Cancer Development. Genes 2021, 12, 269. [Google Scholar] [CrossRef]
- Wang, L.; Shen, H.; Zhu, D.; Feng, B.; Yu, L.; Tian, X.; Ren, C.; Gao, C.; Li, X.; Ma, D.; et al. Increased high mobility group A 2 expression promotes transition of cervical intraepithelial neoplasm into cervical cancer. Oncotarget 2018, 9, 7891–7901. [Google Scholar] [CrossRef] [Green Version]
- Natarajan, S.; Begum, F.; Gim, J.; Wark, L.; Henderson, D.; Davie, J.; Hombach-Klonisch, S.; Klonisch, T. High Mobility Group A2 protects cancer cells against telomere dysfunction. Oncotarget 2016, 7, 12761–12782. [Google Scholar] [CrossRef] [Green Version]
- Di Cello, F.; Hillion, J.; Hristov, A.; Wood, L.J.; Mukherjee, M.; Schuldenfrei, A.; Kowalski, J.; Bhattacharya, R.; Ashfaq, R.; Resar, L.M. HMGA2 participates in transformation in human lung cancer. Mol. Cancer Res. 2008, 6, 743–750. [Google Scholar] [CrossRef] [Green Version]
- El-Telbany, A.; Ma, P.C. Cancer Genes in Lung Cancer: Racial Disparities: Are There Any? Genes Cancer 2012, 3, 467–480. [Google Scholar] [CrossRef]
- Mohammadi, A.; Mansoori, B.; Savadi, P.; Khaze, V.; Minouei, M.; McMillan, N.A.J.; Hallaj-Nezhadi, S.; Baradaran, B. Targeting of high mobility group A2 by small interfering RNA-loaded nanoliposome-induced apoptosis and migration inhibition in gastrointestinal cancer cells. J. Cell. Biochem. 2019, 120, 9203–9212. [Google Scholar] [CrossRef] [PubMed]
- Esmailzadeh, S.; Mansoori, B.; Mohammadi, A.; Shanehbandi, D.; Baradaran, B. siRNA-mediated silencing of HMGA2 induces apoptosis and cell cycle arrest in human colorectal carcinoma. J. Gastrointest. Cancer 2017, 48, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Mansoori, B.; Mohammadi, A.; Asadzadeh, Z.; Shirjang, S.; Minouei, M.; Gaballu, F.A.; Shajari, N.; Kazemi, T.; Gjerstrorff, M.F.; Duijf, P.H.; et al. HMGA2 and Bach-1 cooperate to promote breast cancer cell malignancy. J. Cell. Physiol. 2019, 234, 17714–17726. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Huang, W.; He, Z. Dendrimers as Carriers for siRNA Delivery and Gene Silencing: A Review. Sci. World J. 2013, 2013, 630654. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Chen, Q.; Zheng, Y.; Zhang, P.; Chen, X.; Lu, J.; Lv, Y.; Sun, S.; Zeng, W. PAMAM-cRGD mediating efficient siRNA delivery to spermatogonial stem cells. Stem Cell Res. Ther. 2019, 10, 399. [Google Scholar] [CrossRef] [Green Version]
- Kesharwani, P.; Banerjee, S.; Gupta, U.; Amin, M.C.I.M.; Padhye, S.; Sarkar, F.H.; Iyer, A.K. PAMAM dendrimers as promising nanocarriers for RNAi therapeutics. Mater. Today 2015, 18, 565–572. [Google Scholar] [CrossRef]
- Zarebkohan, A.; Najafi, F.; Moghimi, H.R.; Hemmati, M.; Deevband, M.R.; Kazemi, B. Synthesis and characterization of a PAMAM dendrimer nanocarrier functionalized by SRL peptide for targeted gene delivery to the brain. Eur. J. Pharm. Sci. 2015, 78, 19–30. [Google Scholar] [CrossRef]
- Jensen, L.B.; Pavan, G.M.; Kasimova, M.R.; Rutherford, S.; Danani, A.; Nielsen, H.M.; Foged, C. Elucidating the molecular mechanism of PAMAM–siRNA dendriplex self-assembly: Effect of dendrimer charge density. Int. J. Pharm. 2011, 416, 410–418. [Google Scholar] [CrossRef]
- Hristov, A.C.; Cope, L.; Reyes, M.D.; Singh, M.; Iacobuzio-Donahue, C.; Maitra, A.; Resar, L.M.S. HMGA2 protein expression correlates with lymph node metastasis and increased tumor grade in pancreatic ductal adenocarcinoma. Mod. Pathol. 2009, 22, 43–49. [Google Scholar] [CrossRef] [Green Version]
- Monirinasab, H.; Asadi, H.; Rostamizadeh, K.; Esmaeilzadeh, A.; Khodaei, M.; Fathi, M. Novel lipid-polymer hybrid nanoparticles for siRNA delivery and IGF-1R gene silencing in breast cancer cells. J. Drug Deliv. Sci. Technol. 2018, 48, 96–105. [Google Scholar] [CrossRef]
- Hu, H.; Wang, H.; Liang, S.; Li, X.; Wang, D. Synthesis and characterization of a PAMAM dendrimer nanocarrier functionalized by HA for targeted gene delivery systems and evaluation in vitro. J. Biomater. Sci. Polym. Ed. 2021, 32, 205–228. [Google Scholar] [CrossRef]
- Xu, L.; Yeudall, W.A.; Yang, H. Folic acid-decorated polyamidoamine dendrimer exhibits high tumor uptake and sustained highly localized retention in solid tumors: Its utility for local siRNA delivery. Acta Biomater. 2017, 57, 251–261. [Google Scholar] [CrossRef]
- Rahimi, M.; Shafiei-Irannejad, V.; Safa, K.D.; Salehi, R. Multi-branched ionic liquid-chitosan as a smart and biocompatible nano-vehicle for combination chemotherapy with stealth and targeted properties. Carbohydr. Polym. 2018, 196, 299–312. [Google Scholar] [CrossRef]
- Tanis, I.; Karatasos, K. Association of a Weakly Acidic Anti-Inflammatory Drug (Ibuprofen) with a Poly(Amidoamine) Dendrimer as Studied by Molecular Dynamics Simulations. J. Phys. Chem. B 2009, 113, 10984–10993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, P.; Bandyopadhyay, A.; Chakraborty, A.; Sarkar, K. Enhancement of anticancer activity and drug delivery of chitosan-curcumin nanoparticle via molecular docking and simulation analysis. Carbohydr. Polym. 2018, 182, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, N.; Grenier, P.; Mahmoudi, M.; Lima, E.M.; Appel, E.A.; Dormont, F.; Lim, J.-M.; Karnik, R.; Langer, R.; Farokhzad, O.C. Mechanistic understanding of in vivo protein corona formation on polymeric nanoparticles and impact on pharmacokinetics. Nat. Commun. 2017, 8, 777. [Google Scholar] [CrossRef]
- Lewandowski, W.; Fruhnert, M.; Mieczkowski, J.; Rockstuhl, C.; Górecka, E. Dynamically self-assembled silver nanoparticles as a thermally tunable metamaterial. Nat. Commun. 2015, 6, 6590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, P.T.; Choi, S.K. Mechanisms and implications of dual-acting methotrexate in folate-targeted nanotherapeutic delivery. Int. J. Mol. Sci. 2015, 16, 1772–1790. [Google Scholar] [CrossRef] [Green Version]
- Junyaprasert, V.B.; Dhanahiranpruk, S.; Suksiriworapong, J.; Sripha, K.; Moongkarndi, P. Enhanced toxicity and cellular uptake of methotrexate-conjugated nanoparticles in folate receptor-positive cancer cells by decorating with folic acid-conjugated d-α-tocopheryl polyethylene glycol 1000 succinate. Colloids Surf. B Biointerfaces 2015, 136, 383–393. [Google Scholar] [CrossRef]
- Majoros, I.J.; Williams, C.R.; Becker, A.; Baker, J.R., Jr. Methotrexate delivery via folate targeted dendrimer-based nanotherapeutic platform. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2009, 1, 502–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, R.; Majoros, I.; Misra, A.C.; Koch, A.E.; Campbell, P.; Marotte, H.; Bergin, I.L.; Cao, Z.; Goonewardena, S.; Morry, J.; et al. Folate Receptor-Targeted Dendrimer-Methotrexate Conjugate for Inflammatory Arthritis. J. Biomed. Nanotechnol. 2015, 11, 1431–1441. [Google Scholar] [CrossRef] [PubMed]
- Naghizadeh, S.; Mansoori, B.; Mohammadi, A.; Kafil, H.S.; Mousavi, Z.; Sakhinia, E.; Baradaran, B. Effects of HMGA2 gene downregulation by siRNA on lung carcinoma cell migration in A549 cell lines. J. Cell. Biochem. 2019, 120, 5024–5032. [Google Scholar] [CrossRef] [PubMed]
- SShi, X.; Tian, B.; Ma, W.; Zhang, N.; Qiao, Y.; Li, X.; Zhang, Y.; Huang, B.; Lu, J. A novel anti-proliferative role of HMGA2 in induction of apoptosis through caspase 2 in primary human fibroblast cells. Biosci. Rep. 2015, 35, e00169. [Google Scholar] [CrossRef] [PubMed]
- Meier, P.; Vousden, K.H. Lucifer’s labyrinth—Ten years of path finding in cell death. Mol. Cell 2007, 28, 746–754. [Google Scholar] [CrossRef]
- Zhang, S.; Mo, Q.; Wang, X. Oncological role of HMGA2. Int. J. Oncol. 2019, 5, 775–788. [Google Scholar]
- Mansoori, B.; Mohammadi, A.; Shirjang, S.; Baradaran, B. HMGI-C suppressing induces P53/caspase9 axis to regulate apoptosis in breast adenocarcinoma cells. Cell Cycle 2016, 15, 2585–2592. [Google Scholar] [CrossRef]
- Elango, T.; Thirupathi, A.; Subramanian, S.; Ethiraj, P.; Dayalan, H.; Gnanaraj, P. Methotrexate treatment provokes apoptosis of proliferating keratinocyte in psoriasis patients. Clin. Exp. Med. 2017, 17, 371–381. [Google Scholar] [CrossRef]




| Primers | Sequences | |
|---|---|---|
| β actin | Forward | 5′-TCCCTGGAGAAGAGCTACG-3′ |
| Reverse | 5′-GTAGTTTCGTGGATGCCACA-3′ | |
| HMGA2 | Forward | 5′-TGGGAGGAGCGAAATCTAAA-3′ |
| Reverse | 5′-TCCCTGGAGAAGAGCTACG-3′ | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abedi Gaballu, F.; Cho, W.C.-S.; Dehghan, G.; Zarebkohan, A.; Baradaran, B.; Mansoori, B.; Abbaspour-Ravasjani, S.; Mohammadi, A.; Sheibani, N.; Aghanejad, A.; et al. Silencing of HMGA2 by siRNA Loaded Methotrexate Functionalized Polyamidoamine Dendrimer for Human Breast Cancer Cell Therapy. Genes 2021, 12, 1102. https://0-doi-org.brum.beds.ac.uk/10.3390/genes12071102
Abedi Gaballu F, Cho WC-S, Dehghan G, Zarebkohan A, Baradaran B, Mansoori B, Abbaspour-Ravasjani S, Mohammadi A, Sheibani N, Aghanejad A, et al. Silencing of HMGA2 by siRNA Loaded Methotrexate Functionalized Polyamidoamine Dendrimer for Human Breast Cancer Cell Therapy. Genes. 2021; 12(7):1102. https://0-doi-org.brum.beds.ac.uk/10.3390/genes12071102
Chicago/Turabian StyleAbedi Gaballu, Fereydoon, William Chi-Shing Cho, Gholamreza Dehghan, Amir Zarebkohan, Behzad Baradaran, Behzad Mansoori, Soheil Abbaspour-Ravasjani, Ali Mohammadi, Nader Sheibani, Ayuob Aghanejad, and et al. 2021. "Silencing of HMGA2 by siRNA Loaded Methotrexate Functionalized Polyamidoamine Dendrimer for Human Breast Cancer Cell Therapy" Genes 12, no. 7: 1102. https://0-doi-org.brum.beds.ac.uk/10.3390/genes12071102







