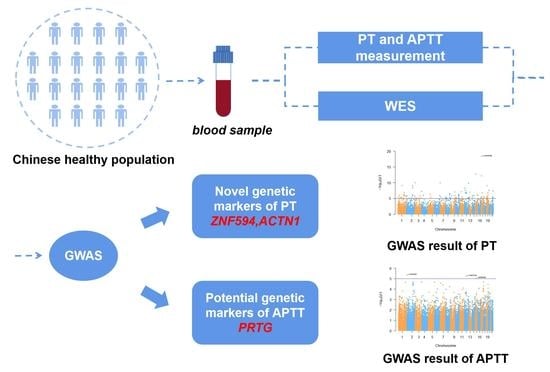
Genetic Polymorphisms Associated with Prothrombin Time and Activated Partial Thromboplastin Time in Chinese Healthy Population
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. PT and APTT Measurement
2.3. Whole-Exome Sequencing and Quality Control
2.4. Data Analysis and Functional Annotation
3. Results
3.1. Baseline Characteristics and Coagulation Parameters
3.2. Discovery of Candidate SNPs for PT and APTT
3.3. Pathway Enrichment and PPI
3.4. Heritability and Phenotypic Variance Explained
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chandler, E.; Kakkar, N.; Kaur, R. Comparison of Rapid Centrifugation Technique with Conventional Centrifugation for Prothrombin Time (PT) and Activated Partial Thromboplastin Time (APTT) Testing. Indian J. Hematol. Blood Transfus. 2019, 35, 161–166. [Google Scholar] [CrossRef]
- Samuelson, B.T.; Cuker, A.; Siegal, D.M.; Crowther, M.; Garcia, D.A. Laboratory Assessment of the Anticoagulant Activity of Direct Oral Anticoagulants: A Systematic Review. Chest 2017, 151, 127–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Favaloro, E.J.; Kershaw, G.; Mohammed, S.; Lippi, G. How to Optimize Activated Partial Thromboplastin Time (APTT) Testing: Solutions to Establishing and Verifying Normal Reference Intervals and Assessing APTT Reagents for Sensitivity to Heparin, Lupus Anticoagulant, and Clotting Factors. Semin. Thromb. Hemost. 2019, 45, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Winter, W.E.; Flax, S.D.; Harris, N.S. Coagulation Testing in the Core Laboratory. Lab. Med. 2017, 48, 295–313. [Google Scholar] [CrossRef] [PubMed]
- Dunois, C. Laboratory Monitoring of Direct Oral Anticoagulants (DOACs). Biomedicines 2021, 9, 445. [Google Scholar] [CrossRef]
- Pipilis, A.; Makrygiannis, S.; Anagnostou, G.; Kaliampakos, S.; Tsakonas, G.; Sourlas, N.; Mallios, P.; Kostelidou, T. Dabigatran plasma levels, aPTT and thromboelastography in patients with AF: Implications for allowing early non-elective surgical procedures. J. Thromb. Thrombolysis 2017, 44, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Achey, M.A.; Nag, U.P.; Robinson, V.L.; Reed, C.R.; Arepally, G.M.; Levy, J.H.; Tracy, E.T. The Developing Balance of Thrombosis and Hemorrhage in Pediatric Surgery: Clinical Implications of Age-Related Changes in Hemostasis. Clin. Appl. Thromb. Hemost. 2020, 26, 1076029620929092. [Google Scholar] [CrossRef]
- Tang, W.; Schwienbacher, C.; Lopez, L.M.; Ben-Shlomo, Y.; Oudot-Mellakh, T.; Johnson, A.D.; Samani, N.J.; Basu, S.; Gögele, M.; Davies, G.; et al. Genetic associations for activated partial thromboplastin time and prothrombin time, their gene expression profiles, and risk of coronary artery disease. Am. J. Hum. Genet. 2012, 91, 152–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weng, L.C.; Cushman, M.; Pankow, J.S.; Basu, S.; Boerwinkle, E.; Folsom, A.R.; Tang, W. A genetic association study of activated partial thromboplastin time in European Americans and African Americans: The ARIC Study. Hum. Mol. Genet. 2015, 24, 2401–2408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, L.; Pu, L.; Ding, Y.; Li, M.; Cabanero, M.; Xie, J.; Zhou, D.; Yang, D.; Zhang, C.; Wang, H.; et al. Relationship between JAK2V617F mutation, allele burden and coagulation function in Ph-negative myeloproliferative neoplasms. Hematology 2017, 22, 354–360. [Google Scholar] [CrossRef]
- Liu, Z.; Xie, Q.; Zhang, H.; Mu, G.; Zhou, S.; Wang, Z.; Jiang, J.; Xiang, Q.; Cui, Y. Target Drug-Calibrated Anti-Xa Activity Assays and Expected Peak-Trough Levels in an Asian Population: A Multicenter Study. Am. J. Cardiovasc. Drugs 2021, 21, 669–679. [Google Scholar] [CrossRef] [PubMed]
- Budkowska, M.; Lebiecka, A.; Marcinowska, Z.; Woźniak, J.; Jastrzębska, M.; Dołęgowska, B. The circadian rhythm of selected parameters of the hemostasis system in healthy people. Thromb. Res. 2019, 182, 79–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warr, A.; Robert, C.; Hume, D.; Archibald, A.; Deeb, N.; Watson, M. Exome Sequencing: Current and Future Perspectives. Genetics 2015, 5, 1543–1550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, M.C.; Haferlach, T.; Nyvold, C.G. A decade with whole exome sequencing in haematology. Br. J. Haematol. 2020, 188, 367–382. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.R-project.org/ (accessed on 11 October 2022).
- Pruim, R.J.; Welch, R.P.; Sanna, S.; Teslovich, T.M.; Chines, P.S.; Gliedt, T.P.; Boehnke, M.; Abecasis, G.R.; Willer, C.J. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics 2010, 26, 2336–2337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, L.D.; Kellis, M. HaploReg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012, 40, D930–D934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Nikpay, M.; Stewart, A.F.R.; McPherson, R. Partitioning the heritability of coronary artery disease highlights the importance of immune-mediated processes and epigenetic sites associated with transcriptional activity. Cardiovasc. Res. 2017, 113, 973–983. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.; Zhou, X. Statistical methods for SNP heritability estimation and partition: A review. Comput. Struct. Biotechnol. J. 2020, 18, 1557–1568. [Google Scholar] [CrossRef]
- Abrink, M.; Aveskogh, M.; Hellman, L. Isolation of cDNA clones for 42 different Krüppel-related zinc finger proteins expressed in the human monoblast cell line U-937. DNA Cell Biol. 1995, 14, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Bazan-Socha, S.; Buregwa-Czuma, S.; Jakiela, B.; Zareba, L.; Zawlik, I.; Myszka, A.; Soja, J.; Okon, K.; Zarychta, J.; Kozlik, P.; et al. Reticular Basement Membrane Thickness is Associated with Growth- and Fibrosis-Promoting Airway Transcriptome Profile-Study in Asthma Patients. Int. J. Mol. Sci. 2021, 22, 998. [Google Scholar] [CrossRef] [PubMed]
- Yan, D.; Hamed, O.; Joshi, T.; Mostafa, M.M.; Jamieson, K.C.; Joshi, R.; Newton, R.; Giembycz, M.A. Analysis of the Indacaterol-Regulated Transcriptome in Human Airway Epithelial Cells Implicates Gene Expression Changes in the Adverse and Therapeutic Effects of β(2)-Adrenoceptor Agonists. J. Pharmacol. Exp. Ther. 2018, 366, 220–236. [Google Scholar] [CrossRef] [Green Version]
- Park, J.A.; Sharif, A.S.; Tschumperlin, D.J.; Lau, L.; Limbrey, R.; Howarth, P.; Drazen, J.M. Tissue factor-bearing exosome secretion from human mechanically stimulated bronchial epithelial cells in vitro and in vivo. J. Allergy Clin. Immunol. 2012, 130, 1375–1383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perrio, M.J.; Ewen, D.; Trevethick, M.A.; Salmon, G.P.; Shute, J.K. Fibrin formation by wounded bronchial epithelial cell layers in vitro is essential for normal epithelial repair and independent of plasma proteins. Clin. Exp. Allergy 2007, 37, 1688–1700. [Google Scholar] [CrossRef] [PubMed]
- Idzko, M.; Pitchford, S.; Page, C. Role of platelets in allergic airway inflammation. J. Allergy Clin. Immunol. 2015, 135, 1416–1423. [Google Scholar] [CrossRef]
- Watson, S.P. Platelet activation by extracellular matrix proteins in haemostasis and thrombosis. Curr. Pharm. Des. 2009, 15, 1358–1372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aslan, J.E.; McCarty, O.J. Rho GTPases in platelet function. J. Thromb. Haemost. 2013, 11, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Sit, S.T.; Manser, E. Rho GTPases and their role in organizing the actin cytoskeleton. J. Cell Sci. 2011, 124, 679–683. [Google Scholar] [CrossRef] [Green Version]
- Delaney, M.K.; Liu, J.; Kim, K.; Shen, B.; Stojanovic-Terpo, A.; Zheng, Y.; Cho, J.; Du, X. Agonist-induced platelet procoagulant activity requires shear and a Rac1-dependent signaling mechanism. Blood 2014, 124, 1957–1967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aslan, J.E.; Tormoen, G.W.; Loren, C.P.; Pang, J.; McCarty, O.J. S6K1 and mTOR regulate Rac1-driven platelet activation and aggregation. Blood 2011, 118, 3129–3136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aslan, J.E.; Baker, S.M.; Loren, C.P.; Haley, K.M.; Itakura, A.; Pang, J.; Greenberg, D.L.; David, L.L.; Manser, E.; Chernoff, J.; et al. The PAK system links Rho GTPase signaling to thrombin-mediated platelet activation. Am. J. Physiol. Cell Physiol. 2013, 305, C519–C528. [Google Scholar] [CrossRef]
- Otey, C.A.; Carpen, O. Alpha-actinin revisited: A fresh look at an old player. Cell Motil. Cytoskelet. 2004, 58, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Sjöblom, B.; Salmazo, A.; Djinović-Carugo, K. Alpha-actinin structure and regulation. Cell Mol. Life Sci. 2008, 65, 2688–2701. [Google Scholar] [CrossRef] [PubMed]
- Schick, U.M.; Jain, D.; Hodonsky, C.J.; Morrison, J.V.; Davis, J.P.; Brown, L.; Sofer, T.; Conomos, M.P.; Schurmann, C.; McHugh, C.P.; et al. Genome-wide Association Study of Platelet Count Identifies Ancestry-Specific Loci in Hispanic/Latino Americans. Am. J. Hum. Genet. 2016, 98, 229–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kunishima, S.; Okuno, Y.; Yoshida, K.; Shiraishi, Y.; Sanada, M.; Muramatsu, H.; Chiba, K.; Tanaka, H.; Miyazaki, K.; Sakai, M.; et al. ACTN1 mutations cause congenital macrothrombocytopenia. Am. J. Hum. Genet. 2013, 92, 431–438. [Google Scholar] [CrossRef] [Green Version]
- Willems, E.; Dedobbeleer, M.; Digregorio, M.; Lombard, A.; Lumapat, P.N.; Rogister, B. The functional diversity of Aurora kinases: A comprehensive review. Cell Div. 2018, 13, 7. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, H.G.; Yu, G.; Makitalo, M.; Yang, D.; Xie, H.X.; Jones, M.R.; Ravid, K. Conditional overexpression of transgenes in megakaryocytes and platelets in vivo. Blood 2005, 106, 1559–1564. [Google Scholar] [CrossRef] [PubMed]
- Lordier, L.; Chang, Y.; Jalil, A.; Aurade, F.; Garçon, L.; Lécluse, Y.; Larbret, F.; Kawashima, T.; Kitamura, T.; Larghero, J.; et al. Aurora B is dispensable for megakaryocyte polyploidization, but contributes to the endomitotic process. Blood 2010, 116, 2345–2355. [Google Scholar] [CrossRef] [PubMed]
- Tovuu, L.O.; Utsunomiya, T.; Imura, S.; Morine, Y.; Ikemoto, T.; Arakawa, Y.; Mori, H.; Hanaoka, J.; Kanamoto, M.; Sugimoto, K.; et al. The role of Aurora B expression in non-tumor liver tissues of patients with hepatocellular carcinoma. Int. J. Clin. Oncol. 2014, 19, 622–628. [Google Scholar] [CrossRef] [PubMed]
- Eicher, J.D.; Wakabayashi, Y.; Vitseva, O.; Esa, N.; Yang, Y.; Zhu, J.; Freedman, J.E.; McManus, D.D.; Johnson, A.D. Characterization of the platelet transcriptome by RNA sequencing in patients with acute myocardial infarction. Platelets 2016, 27, 230–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricklin, D.; Reis, E.S.; Lambris, J.D. Complement in disease: A defence system turning offensive. Nat. Rev. Nephrol. 2016, 12, 383–401. [Google Scholar] [CrossRef] [Green Version]
- Subramaniam, S.; Jurk, K.; Hobohm, L.; Jäckel, S.; Saffarzadeh, M.; Schwierczek, K.; Wenzel, P.; Langer, F.; Reinhardt, C.; Ruf, W. Distinct contributions of complement factors to platelet activation and fibrin formation in venous thrombus development. Blood 2017, 129, 2291–2302. [Google Scholar] [CrossRef] [Green Version]
- Vlaicu, S.I.; Tatomir, A.; Rus, V.; Rus, H. Role of C5b-9 and RGC-32 in Cancer. Front. Immunol. 2019, 10, 1054. [Google Scholar] [CrossRef] [Green Version]
- Wiedmer, T.; Esmon, C.T.; Sims, P.J. On the mechanism by which complement proteins C5b-9 increase platelet prothrombinase activity. J. Biol. Chem. 1986, 261, 14587–14592. [Google Scholar] [CrossRef]
- Rudic, R.D.; McNamara, P.; Reilly, D.; Grosser, T.; Curtis, A.M.; Price, T.S.; Panda, S.; Hogenesch, J.B.; FitzGerald, G.A. Bioinformatic analysis of circadian gene oscillation in mouse aorta. Circulation 2005, 112, 2716–2724. [Google Scholar] [CrossRef] [Green Version]
- Takeda, N.; Maemura, K.; Horie, S.; Oishi, K.; Imai, Y.; Harada, T.; Saito, T.; Shiga, T.; Amiya, E.; Manabe, I.; et al. Thrombomodulin is a clock-controlled gene in vascular endothelial cells. J. Biol. Chem. 2007, 282, 32561–32567. [Google Scholar] [CrossRef] [Green Version]
- Cheng, S.; Jiang, Z.; Zou, Y.; Chen, C.; Wang, Y.; Liu, Y.; Xiao, J.; Guo, H.; Wang, Z. Downregulation of Clock in circulatory system leads to an enhancement of fibrinolysis in mice. Exp. Biol. Med. 2011, 236, 1078–1084. [Google Scholar] [CrossRef]
- Ohkura, N.; Oishi, K.; Fukushima, N.; Kasamatsu, M.; Atsumi, G.I.; Ishida, N.; Horie, S.; Matsuda, J. Circadian clock molecules CLOCK and CRYs modulate fibrinolytic activity by regulating the PAI-1 gene expression. J. Thromb. Haemost. 2006, 4, 2478–2485. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, R.; Mauer, A.C.; Lino Cardenas, C.L.; Guo, X.; Yao, J.; Zhang, X.; Wunderer, F.; Smith, A.V.; Wong, Q.; Pechlivanis, S.; et al. HDAC9 is implicated in atherosclerotic aortic calcification and affects vascular smooth muscle cell phenotype. Nat. Genet. 2019, 51, 1580–1587. [Google Scholar] [CrossRef]
- Kuang, X.; Chen, S.; Lao, J.; Chen, Y.; Jia, D.; Tu, L.; Ma, L.; Liao, X.; Zhao, W.; Li, Q. HDAC9 in the Injury of Vascular Endothelial Cell Mediated by P38 MAPK Pathway. J. Interferon Cytokine Res. 2021, 41, 439–449. [Google Scholar] [CrossRef] [PubMed]
- Danckwardt, S.; Gantzert, A.S.; Macher-Goeppinger, S.; Probst, H.C.; Gentzel, M.; Wilm, M.; Gröne, H.J.; Schirmacher, P.; Hentze, M.W.; Kulozik, A.E. p38 MAPK controls prothrombin expression by regulated RNA 3′ end processing. Mol. Cell 2011, 41, 298–310. [Google Scholar] [CrossRef]
- Branger, J.; van den Blink, B.; Weijer, S.; Gupta, A.; van Deventer, S.J.; Hack, C.E.; Peppelenbosch, M.P.; van der Poll, T. Inhibition of coagulation, fibrinolysis, and endothelial cell activation by a p38 mitogen-activated protein kinase inhibitor during human endotoxemia. Blood 2003, 101, 4446–4448. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, T.; Subramanian, A.; Albert, V.; Kumar, A.; Prakash, S.; Pati, H.P. Platelet Function Analysis by Flowcytometry in Thrombocytopenic Trauma Patients. Indian J. Hematol. Blood Transfus. 2021, 37, 398–403. [Google Scholar] [CrossRef]
- Xiang, T.; Yuan, C.; Guo, X.; Wang, H.; Cai, Q.; Xiang, Y.; Luo, W.; Liu, G. The novel ZEB1-upregulated protein PRTG induced by Helicobacter pylori infection promotes gastric carcinogenesis through the cGMP/PKG signaling pathway. Cell Death Dis. 2021, 12, 150. [Google Scholar] [CrossRef] [PubMed]
- Aburima, A.; Walladbegi, K.; Wake, J.D.; Naseem, K.M. cGMP signaling inhibits platelet shape change through regulation of the RhoA-Rho Kinase-MLC phosphatase signaling pathway. J. Thromb. Haemost. 2017, 15, 1668–1678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Offermanns, S. Activation of platelet function through G protein-coupled receptors. Circ. Res. 2006, 99, 1293–1304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Qian, L.; Liu, Y.; Liu, Y.; Yu, W.; Zhao, Y. CircRNA-ceRNA Network Revealing the Potential Regulatory Roles of CircRNA in Alzheimer’s Disease Involved the cGMP-PKG Signal Pathway. Front. Mol. Neurosci. 2021, 14, 665788. [Google Scholar] [CrossRef] [PubMed]
- Lopes-Pires, M.E.; Naime, A.C.; Almeida Cardelli, N.J.; Anjos, D.J.; Antunes, E.; Marcondes, S. PKC and AKT Modulate cGMP/PKG Signaling Pathway on Platelet Aggregation in Experimental Sepsis. PLoS ONE 2015, 10, e0137901. [Google Scholar] [CrossRef] [PubMed]
- Makhoul, S.; Trabold, K.; Gambaryan, S.; Tenzer, S.; Pillitteri, D.; Walter, U.; Jurk, K. cAMP- and cGMP-elevating agents inhibit GPIbα-mediated aggregation but not GPIbα-stimulated Syk activation in human platelets. Cell Commun. Signal. 2019, 17, 122. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Zhou, M.; Dong, A.; Loppnau, P.; Wang, M.; Min, J.; Liu, K. Structural basis of the TAM domain of BAZ2A in binding to DNA or RNA independent of methylation status. J. Biol. Chem. 2021, 297, 101351. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Frommel, S.C.; Oakes, C.C.; Simon, R.; Grupp, K.; Gerig, C.Y.; Bär, D.; Robinson, M.D.; Baer, C.; Weiss, M.; et al. BAZ2A (TIP5) is involved in epigenetic alterations in prostate cancer and its overexpression predicts disease recurrence. Nat. Genet. 2015, 47, 22–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Wu, W.; Ding, H.; Li, Q.; Xie, K. The transcription factor 7 like 2-binding protein TIP5 activates β-catenin/transcription factor signaling in hepatocellular carcinoma. Mol. Med. Rep. 2018, 17, 7645–7651. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, J.; Zhou, L.; Yin, C. LINC00885 promotes cervical cancer progression through sponging miR-3150b-3p and upregulating BAZ2A. Biol. Direct 2022, 17, 4. [Google Scholar] [CrossRef]
- Hanlon, K.; Rudin, C.E.; Harries, L.W. Investigating the targets of MIR-15a and MIR-16-1 in patients with chronic lymphocytic leukemia (CLL). PLoS ONE 2009, 4, e7169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doench, J.G.; Sharp, P.A. Specificity of microRNA target selection in translational repression. Genes Dev. 2004, 18, 504–511. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Sun, P.; Zhang, K.; Hassan, S.H.; Zhang, X.; Tang, X.; Pu, H.; Stetler, R.A.; Chen, J.; Yin, K.J. Endothelium-Targeted Deletion of microRNA-15a/16-1 Promotes Poststroke Angiogenesis and Improves Long-Term Neurological Recovery. Circ. Res. 2020, 126, 1040–1057. [Google Scholar] [CrossRef] [PubMed]
- Vila, L.; Martinez-Perez, A.; Camacho, M.; Buil, A.; Alcolea, S.; Pujol-Moix, N.; Soler, M.; Antón, R.; Souto, J.C.; Fontcuberta, J.; et al. Heritability of thromboxane A2 and prostaglandin E2 biosynthetic machinery in a Spanish population. Arter. Thromb Vasc. Biol. 2010, 30, 128–134. [Google Scholar] [CrossRef] [Green Version]
- Warren, D.M.; Soria, J.M.; Souto, J.C.; Comuzzie, A.; Fontcuberta, J.; Blangero, J.; MacCluer, J.W.; Almasy, L. Heritability of hemostasis phenotypes and their correlation with type 2 diabetes status in Mexican Americans. Hum. Biol. 2005, 77, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Houlihan, L.M.; Davies, G.; Tenesa, A.; Harris, S.E.; Luciano, M.; Gow, A.J.; McGhee, K.A.; Liewald, D.C.; Porteous, D.J.; Starr, J.M.; et al. Common variants of large effect in F12, KNG1, and HRG are associated with activated partial thromboplastin time. Am. J. Hum. Genet. 2010, 86, 626–631. [Google Scholar] [CrossRef]



| Characteristics | Total |
|---|---|
| N | 403 |
| Age (years) | 29.5 ± 8.8 |
| Age range (years) | 18–60 |
| Female (n) | 133 (33.0%) |
| BMI (kg/m2) | 22.6 ± 1.8 |
| PT (s) | 11.6 ± 1.4 |
| Median PT [range] (s) | 11.5 [9.7, 27.8] |
| APTT (s) | 29.6 ± 5.2 |
| Median APTT [range] (s) | 28.7 [17.8, 46.4] |
| Heritability of PT | Heritability of APTT | ||||
|---|---|---|---|---|---|
| Source | Variance | SE | Source | Variance | SE |
| Vg | 1.57 | 0.78 | Vg | 17.33 | 10.69 |
| Ve | 0.31 | 0.76 | Ve | 9.78 | 10.50 |
| Vp | 1.89 | 0.13 | Vp | 27.11 | 1.92 |
| Vg/Vp | 0.83 | 0.40 | Vg/Vp | 0.64 | 0.39 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, F.; Mu, G.; Liu, Z.; Xie, Q.; Zhang, H.; Zhou, S.; Wang, Z.; Hu, K.; Wang, Z.; Zhao, X.; et al. Genetic Polymorphisms Associated with Prothrombin Time and Activated Partial Thromboplastin Time in Chinese Healthy Population. Genes 2022, 13, 1867. https://0-doi-org.brum.beds.ac.uk/10.3390/genes13101867
Zhang F, Mu G, Liu Z, Xie Q, Zhang H, Zhou S, Wang Z, Hu K, Wang Z, Zhao X, et al. Genetic Polymorphisms Associated with Prothrombin Time and Activated Partial Thromboplastin Time in Chinese Healthy Population. Genes. 2022; 13(10):1867. https://0-doi-org.brum.beds.ac.uk/10.3390/genes13101867
Chicago/Turabian StyleZhang, Fan, Guangyan Mu, Zhiyan Liu, Qiufen Xie, Hanxu Zhang, Shuang Zhou, Zhe Wang, Kun Hu, Zining Wang, Xia Zhao, and et al. 2022. "Genetic Polymorphisms Associated with Prothrombin Time and Activated Partial Thromboplastin Time in Chinese Healthy Population" Genes 13, no. 10: 1867. https://0-doi-org.brum.beds.ac.uk/10.3390/genes13101867