PCYT1A Missense Variant in Vizslas with Disproportionate Dwarfism
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Clinical Examination
2.3. Diagnostic Imaging
2.4. DNA Extraction, Linkage Analysis and Homozygosity Mapping
2.5. Whole-Genome Sequencing
2.6. Variant Calling
2.7. Gene Analysis
2.8. PCR and Sanger Sequencing
3. Results
3.1. Family Anamnesis and Clinical Examinations
3.2. Diagnostic Imaging
3.3. Genetic Analyses
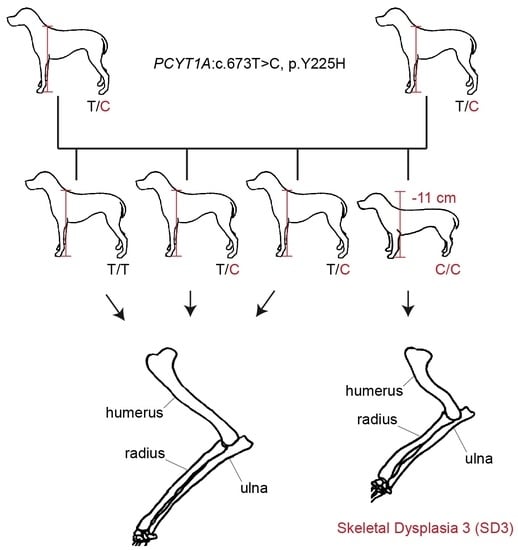
3.4. Genotype–Phenotype Correlation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Krakow, D. Skeletal Dysplasias. Clin. Perinatol. 2015, 42, 301–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mortier, G.R.; Cohn, D.H.; Cormier-Daire, V.; Hall, C.; Krakow, D.; Mundlos, S.; Nishimura, G.; Robertson, S.; Sangiorgi, L.; Savarirayan, R.; et al. Nosology and Classification of Genetic Skeletal Disorders: 2019 Revision. Am. J. Med. Genet. Part A 2019, 179, 2393–2419. [Google Scholar] [CrossRef] [PubMed]
- Sewell, M.D.; Chahal, A.; Al-Hadithy, N.; Blunn, G.W.; Molloy, S.; Hashemi-Nejad, A. Genetic Skeletal Dysplasias: A Guide to Diagnosis and Management. J. Back Musculoskelet. Rehabil. 2015, 28, 575–590. [Google Scholar] [CrossRef]
- Bannasch, D.L.; Baes, C.F.; Leeb, T. Genetic Variants Affecting Skeletal Morphology in Domestic Dogs. Trends Genet. 2020, 36, 598–609. [Google Scholar] [CrossRef] [PubMed]
- Struck, A.-K.; Braun, M.; Detering, K.A.; Dziallas, P.; Neßler, J.; Fehr, M.; Metzger, J.; Distl, O. A Structural UGDH Variant Associated with Standard Munchkin Cats. BMC Genet. 2020, 21, 67. [Google Scholar] [CrossRef]
- Cavanagh, J.A.L.; Tammen, I.; Windsor, P.A.; Bateman, J.F.; Savarirayan, R.; Nicholas, F.W.; Raadsma, H.W. Bulldog Dwarfism in Dexter Cattle Is Caused by Mutations in ACAN. Mamm. Genome 2007, 18, 808–814. [Google Scholar] [CrossRef]
- Rafati, N.; Andersson, L.S.; Mikko, S.; Feng, C.; Raudsepp, T.; Pettersson, J.; Janecka, J.; Wattle, O.; Ameur, A.; Thyreen, G.; et al. Large Deletions at the SHOX Locus in the Pseudoautosomal Region Are Associated with Skeletal Atavism in Shetland Ponies. G3 Genes|Genomes|Genet. 2016, 6, 2213–2223. [Google Scholar] [CrossRef] [Green Version]
- Parker, H.G.; VonHoldt, B.M.; Quignon, P.; Margulies, E.H.; Shao, S.; Mosher, D.S.; Spady, T.C.; Elkahloun, A.; Cargill, M.; Jones, P.G.; et al. An Expressed Fgf4 Retrogene Is Associated with Breed-Defining Chondrodysplasia in Domestic Dogs. Science 2009, 325, 995–998. [Google Scholar] [CrossRef] [Green Version]
- Batcher, K.; Dickinson, P.; Giuffrida, M.; Sturges, B.; Vernau, K.; Knipe, M.; Rasouliha, S.H.; Drögemüller, C.; Leeb, T.; Maciejczyk, K.; et al. Phenotypic Effects of FGF4 Retrogenes on Intervertebral Disc Disease in Dogs. Genes 2019, 10, 435. [Google Scholar] [CrossRef] [Green Version]
- Brown, E.A.; Dickinson, P.J.; Mansour, T.; Sturges, B.K.; Aguilar, M.; Young, A.E.; Korff, C.; Lind, J.; Ettinger, C.L.; Varon, S.; et al. FGF4 Retrogene on CFA12 Is Responsible for Chondrodystrophy and Intervertebral Disc Disease in Dogs. Proc. Natl. Acad. Sci. USA 2017, 114, 11476–11481. [Google Scholar] [CrossRef]
- Rudd Garces, G.; Turba, M.E.; Muracchini, M.; Diana, A.; Jagannathan, V.; Gentilini, F.; Leeb, T. PRKG2 Splice Site Variant in Dogo Argentino Dogs with Disproportionate Dwarfism. Genes 2021, 12, 1489. [Google Scholar] [CrossRef]
- Frischknecht, M.; Niehof-Oellers, H.; Jagannathan, V.; Owczarek-Lipska, M.; Drögemüller, C.; Dietschi, E.; Dolf, G.; Tellhelm, B.; Lang, J.; Tiira, K.; et al. A COL11A2 Mutation in Labrador Retrievers with Mild Disproportionate Dwarfism. PLoS ONE 2013, 8, e60149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kyöstilä, K.; Lappalainen, A.K.; Lohi, H. Canine Chondrodysplasia Caused by a Truncating Mutation in Collagen-Binding Integrin Alpha Subunit 10. PLoS ONE 2013, 8, e75621. [Google Scholar] [CrossRef] [Green Version]
- Neff, M.W.; Beck, J.S.; Koeman, J.M.; Boguslawski, E.; Kefene, L.; Borgman, A.; Ruhe, A.L. Partial Deletion of the Sulfate Transporter SLC13A1 Is Associated with an Osteochondrodysplasia in the Miniature Poodle Breed. PLoS ONE 2012, 7, e51917. [Google Scholar] [CrossRef]
- Goldstein, O.; Guyon, R.; Kukekova, A.; Kuznetsova, T.N.; Pearce-Kelling, S.E.; Johnson, J.; Aguirre, G.D.; Acland, G.M. COL9A2 and COL9A3 Mutations in Canine Autosomal Recessive Oculoskeletal Dysplasia. Mamm. Genome 2010, 21, 398–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stavinohova, R.; Hartley, C.; Burmeister, L.M.; Ricketts, S.L.; Pettitt, L.; Pont, R.T.; Hitti, R.J.; Schofield, E.; Oliver, J.A.C.; Mellersh, C.S. Clinical, Histopathological and Genetic Characterisation of Oculoskeletal Dysplasia in the Northern Inuit Dog. PLoS ONE 2019, 14, e0220761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voorbij, A.M.W.Y.; van Steenbeek, F.G.; Vos-Loohuis, M.; Martens, E.E.C.P.; Hanson-Nilsson, J.M.; van Oost, B.A.; Kooistra, H.S.; Leegwater, P.A. A Contracted DNA Repeat in LHX3 Intron 5 Is Associated with Aberrant Splicing and Pituitary Dwarfism in German Shepherd Dogs. PLoS ONE 2011, 6, e27940. [Google Scholar] [CrossRef]
- Voorbij, A.M.W.Y.; Leegwater, P.A.; Kooistra, H.S. Pituitary Dwarfism in Saarloos and Czechoslovakian Wolfdogs Is Associated with a Mutation in LHX3. J. Vet. Intern. Med. 2014, 28, 1770–1774. [Google Scholar] [CrossRef] [Green Version]
- Thaiwong, T.; Corner, S.; La Forge, S.; Kiupel, M. Dwarfism in Tibetan Terrier Dogs with an LHX3 Mutation. J. Vet. Diagn. Investig. 2021, 33, 740–743. [Google Scholar] [CrossRef]
- Kyöstilä, K.; Niskanen, J.E.; Arumilli, M.; Donner, J.; Hytönen, M.K.; Lohi, H. Intronic Variant in POU1F1 Associated with Canine Pituitary Dwarfism. Hum. Genet. 2021, 140, 1553–1562. [Google Scholar] [CrossRef]
- Iio, A.; Maeda, S.; Yonezawa, T.; Momoi, Y.; Motegi, T. Isolated Growth Hormone Deficiency in a Chihuahua with a GH1 Mutation. J. Vet. Diagn. Investig. 2020, 32, 733–736. [Google Scholar] [CrossRef] [PubMed]
- American Kennel Club. Available online: https://www.akc.org/dog-breeds/vizsla/ (accessed on 10 October 2022).
- Abecasis, G.R.; Cherny, S.S.; Cookson, W.O.; Cardon, L.R. Merlin—Rapid Analysis of Dense Genetic Maps Using Sparse Gene Flow Trees. Nat. Genet. 2002, 30, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jagannathan, V.; Drögemüller, C.; Leeb, T.; Aguirre, G.; André, C.; Bannasch, D.; Becker, D.; Davis, B.; Ekenstedt, K.; Faller, K.; et al. A Comprehensive Biomedical Variant Catalogue Based on Whole Genome Sequences of 582 Dogs and Eight Wolves. Anim. Genet. 2019, 50, 695–704. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and Accurate Long-Read Alignment with Burrows–Wheeler Transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [Green Version]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce Framework for Analyzing next-Generation DNA Sequencing Data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [Green Version]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A Program for Annotating and Predicting the Effects of Single Nucleotide Polymorphisms, SnpEff. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [Green Version]
- Kent, C. Regulatory Enzymes of Phosphatidylcholine Biosynthesis: A Personal Perspective. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids 2005, 1733, 53–66. [Google Scholar] [CrossRef]
- Yamamoto, G.L.; Baratela, W.A.R.; Almeida, T.F.; Lazar, M.; Afonso, C.L.; Oyamada, M.K.; Suzuki, L.; Oliveira, L.A.N.; Ramos, E.S.; Kim, C.A.; et al. Mutations in PCYT1A Cause Spondylometaphyseal Dysplasia with Cone-Rod Dystrophy. Am. J. Hum. Genet. 2014, 94, 113–119. [Google Scholar] [CrossRef]
- Zinser, E.; Sperka-Gottlieb, C.D.M.; Fasch, E.V.; Kohlwein, S.D.; Paltauf, F.; Daum, G. Phospholipid Synthesis and Lipid Composition of Subcellular Membranes in the Unicellular Eukaryote Saccharomyces Cerevisiae. J. Bacteriol. 1991, 173, 2026–2034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Johnson, J.; Ding, Z.; Paetzel, M.; Cornell, R.B. Crystal Structure of a Mammalian CTP: Phosphocholine Cytidylyltransferase Catalytic Domain Reveals Novel Active Site Residues within a Highly Conserved Nucleotidyltransferase Fold. J. Biol. Chem. 2009, 284, 33535–33548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taneva, S.; Dennis, M.K.; Ding, Z.; Smith, J.L.; Cornell, R.B. Contribution of Each Membrane Binding Domain of the CTP:Phosphocholine Cytidylyltransferase-α Dimer to Its Activation, Membrane Binding, and Membrane Cross-Bridging. J. Biol. Chem. 2008, 283, 28137–28148. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Taneva, S.G.; Holland, B.W.; Tieleman, D.P.; Cornell, R.B. Structural Basis for Autoinhibition of CTP:Phosphocholine Cytidylyltransferase (CCT), the Regulatory Enzyme in Phosphatidylcholine Synthesis, by Its Membrane-Binding Amphipathic Helix. J. Biol. Chem. 2014, 289, 1742–1755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wuthier, R.E. Lipids of Matrix Vesicles. Fed. Proc. 1976, 35, 117–121. [Google Scholar] [PubMed]
- Boyan, B.D.; Asmussen, N.C.; Lin, Z.; Schwartz, Z. The Role of Matrix-Bound Extracellular Vesicles in the Regulation of Endochondral Bone Formation. Cells 2022, 11, 1619. [Google Scholar] [CrossRef]
- Hoover-Fong, J.; Sobreira, N.; Jurgens, J.; Modaff, P.; Blout, C.; Moser, A.; Kim, O.-H.; Cho, T.-J.; Cho, S.Y.; Kim, S.J.; et al. Mutations in PCYT1A, Encoding a Key Regulator of Phosphatidylcholine Metabolism, Cause Spondylometaphyseal Dysplasia with Cone-Rod Dystrophy. Am. J. Hum. Genet. 2014, 94, 105–112. [Google Scholar] [CrossRef] [Green Version]
- Payne, F.; Lim, K.; Girousse, A.; Brown, R.J.; Kory, N.; Robbins, A.; Xue, Y.; Sleigh, A.; Cochran, E.; Adams, C.; et al. Mutations Disrupting the Kennedy Phosphatidylcholine Pathway in Humans with Congenital Lipodystrophy and Fatty Liver Disease. Proc. Natl. Acad. Sci. USA 2014, 111, 8901–8906. [Google Scholar] [CrossRef] [Green Version]
- Testa, F.; Filippelli, M.; Brunetti-Pierri, R.; Di Fruscio, G.; Di Iorio, V.; Pizzo, M.; Torella, A.; Barillari, M.R.; Nigro, V.; Brunetti-Pierri, N.; et al. Mutations in the PCYT1A Gene Are Responsible for Isolated Forms of Retinal Dystrophy. Eur. J. Hum. Genet. 2017, 25, 651–655. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Magdaleno, S.; Tabas, I.; Jackowski, S. Early Embryonic Lethality in Mice with Targeted Deletion of the CTP:Phosphocholine Cytidylyltransferase α Gene (Pcyt1a). Mol. Cell. Biol. 2005, 25, 3357–3363. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]





| Filtering Step | Homozygous Variants |
|---|---|
| Variants in the affected Vizsla | 2,905,167 |
| Private variants | 1193 |
| Private protein-changing variants | 8 |
| Private protein-changing variants in critical intervals | 1 |
| Phenotype | T/T | T/C | C/C |
|---|---|---|---|
| Cases (n = 8) | 1 1 | 0 | 7 |
| Obligate carriers (n = 8) | 1 2 | 7 | 0 |
| Unaffected 1st degree relatives (n = 20) | 12 | 8 | 0 |
| Other controls (n = 95) | 84 | 11 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ludwig-Peisker, O.; Ansel, E.; Schweizer, D.; Jagannathan, V.; Loechel, R.; Leeb, T. PCYT1A Missense Variant in Vizslas with Disproportionate Dwarfism. Genes 2022, 13, 2354. https://0-doi-org.brum.beds.ac.uk/10.3390/genes13122354
Ludwig-Peisker O, Ansel E, Schweizer D, Jagannathan V, Loechel R, Leeb T. PCYT1A Missense Variant in Vizslas with Disproportionate Dwarfism. Genes. 2022; 13(12):2354. https://0-doi-org.brum.beds.ac.uk/10.3390/genes13122354
Chicago/Turabian StyleLudwig-Peisker, Odette, Emily Ansel, Daniela Schweizer, Vidhya Jagannathan, Robert Loechel, and Tosso Leeb. 2022. "PCYT1A Missense Variant in Vizslas with Disproportionate Dwarfism" Genes 13, no. 12: 2354. https://0-doi-org.brum.beds.ac.uk/10.3390/genes13122354