Genome-Wide Identification and Structural Analysis of bZIP Transcription Factor Genes in Brassica napus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of bZIP Family Genes in Brassica napus
2.2. Multiple Sequence Alignment and Phylogenetic Analysis of the bZIP Family Members in Brassica napus
2.3. Analysis of Exon-Intron Structures and Conserved Motifs in BnbZIP Genes
2.4. Chromosomal Distribution of BnbZIP Genes in Brassica napus
2.5. Analysis of BnbZIP Gene Expression Patterns
2.6. Quantitative Real Time PCR Validation of Transcriptome Data
3. Results
3.1. Identification of bZIP Family Members in Brassica napus
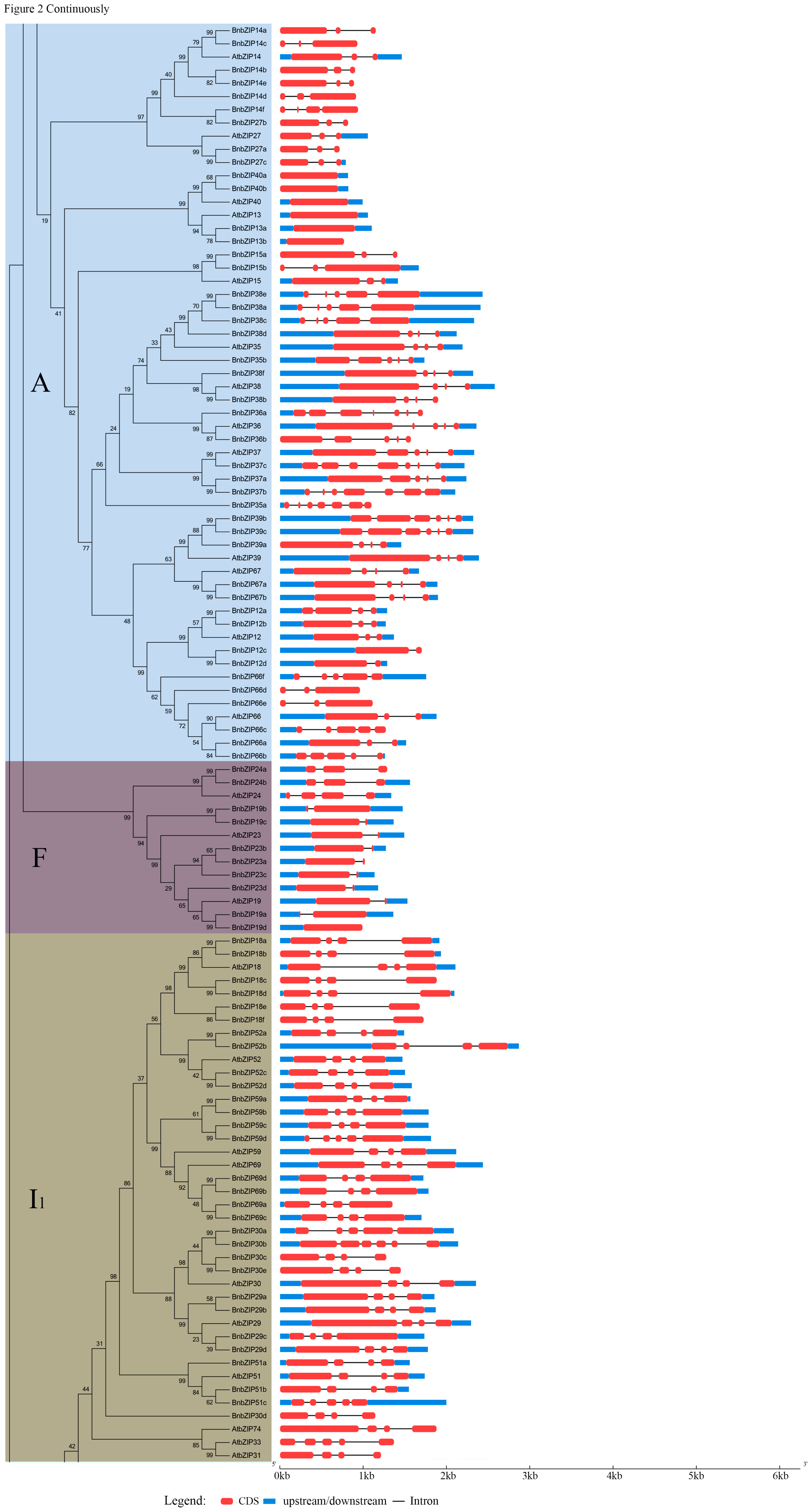
3.2. Phylogenetic Analysis of the bZIP TFs in Brassica napus
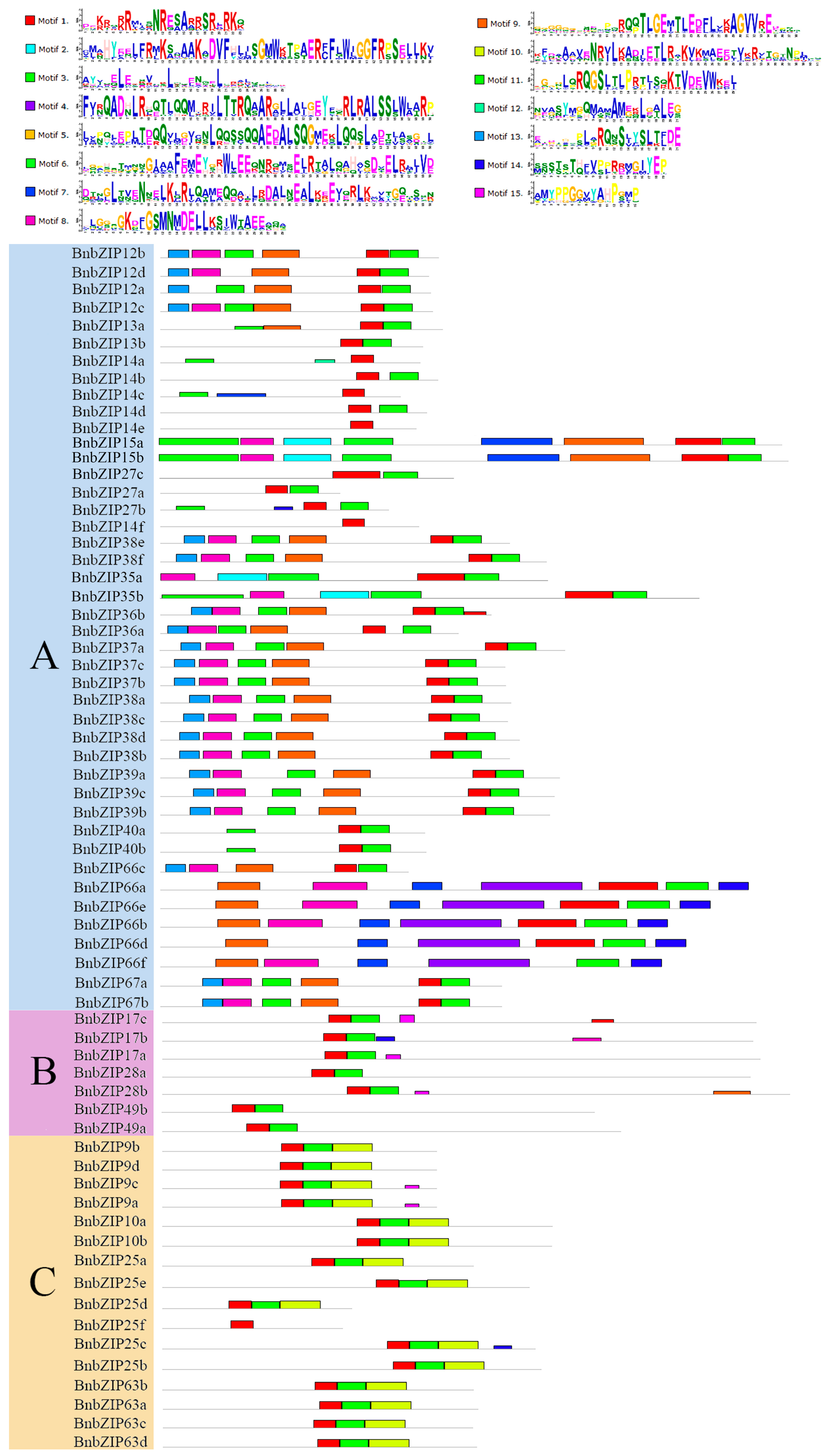
3.3. Gene Structure and Motif Analyses of BnbZIPs
3.4. Genomic Distribution of the 247 BnbZIPs on the 19 Brassica napus Chromosomes
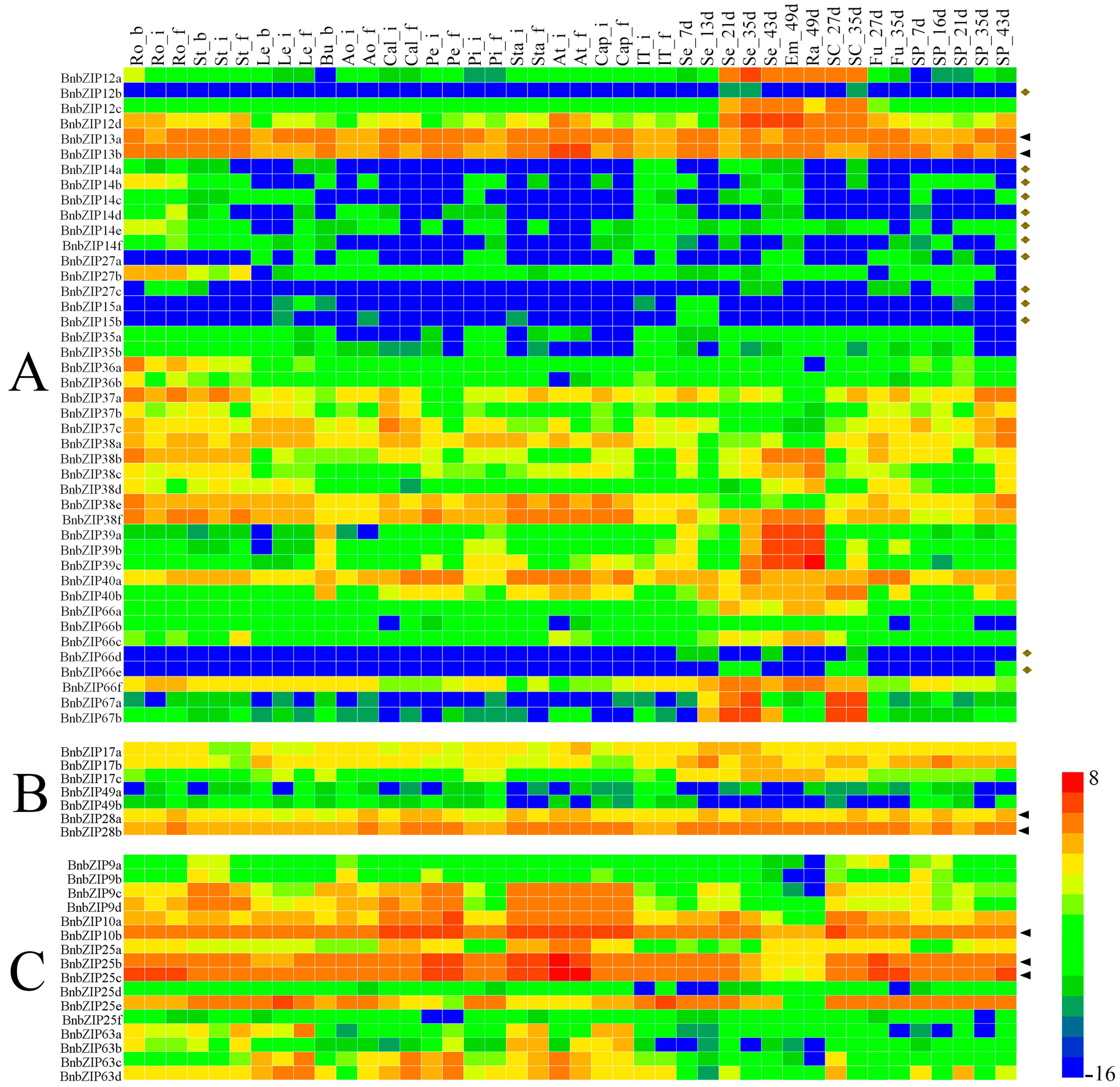
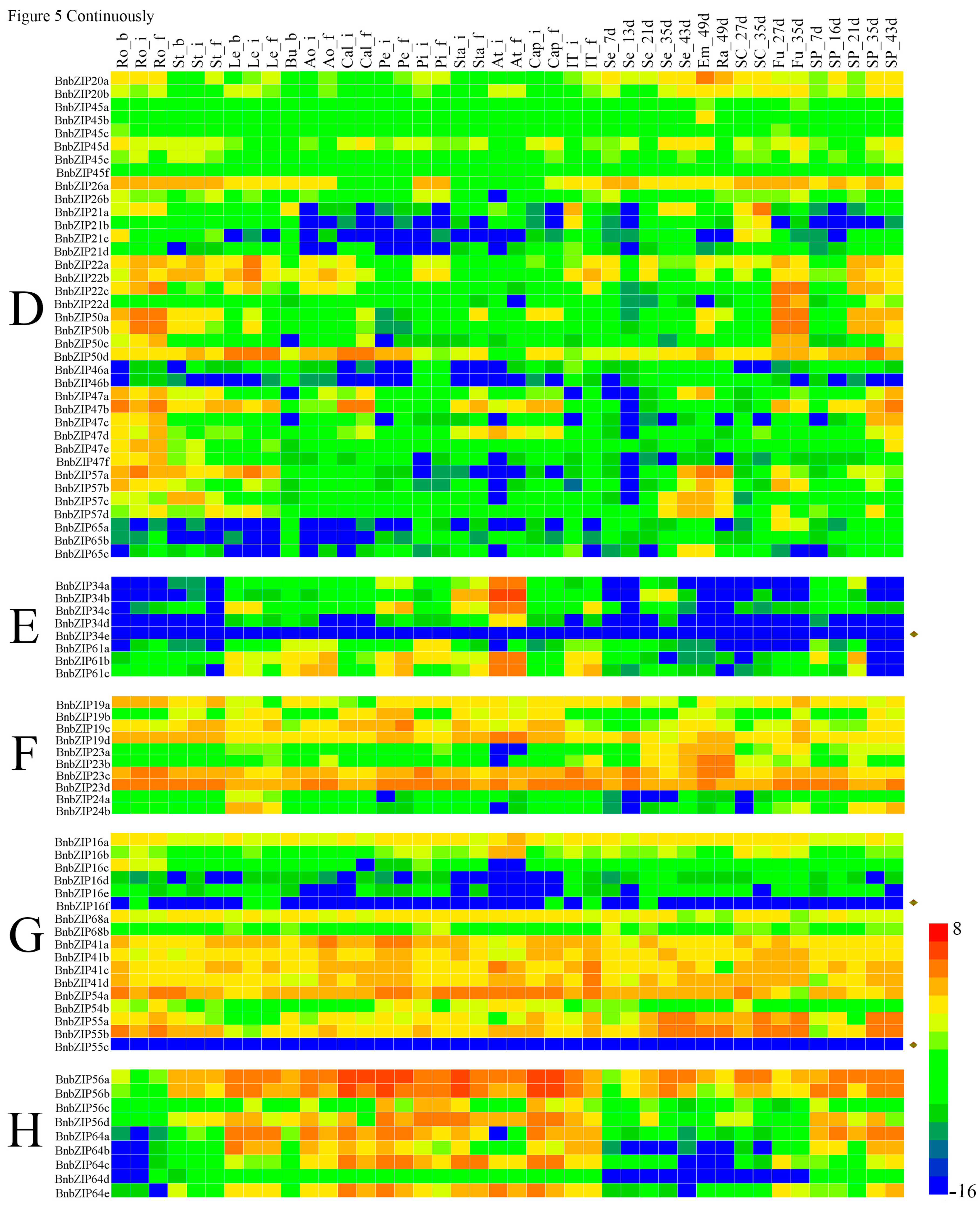
3.5. The Differential Expression Analyses of the BnbZIP Genes in Various Tissues of Brassica napus
4. Discussion
4.1. Characterization of bZIP Gene Families in Brassica napus
4.2. Expression Profile Analysis Reveals the Potential Roles of bZIP Genes in Brassica napus
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Nijhawan, A.; Jain, M.; Tyagi, A.K.; Khurana, J.P. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008, 146, 333–350. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.; Chen, J.; Wang, Y.; Chen, Y.; Chen, S.; Lin, Y.; Pan, S.; Zhong, X.; Xie, D. Genome-Wide Analysis of bZIP-Encoding Genes in Maize. DNA Res. 2012, 19, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R.R.; Lynch, T.J. The Arabidopsis Abscisic Acid Response Gene ABI5 Encodes a Basic Leucine Zipper Transcription Factor. Plant Cell 2000, 12, 599–609. [Google Scholar] [CrossRef] [PubMed]
- Bensmihen, S.; Rippa, S.; Lambert, G.; Jublot, D.; Pautot, V.; Granier, F.; Giraudat, J.; Parcy, F. The Homologous ABI5 and EEL Transcription Factors Function Antagonistically to Fine-Tune Gene Expression during Late Embryogenesis. Plant Cell 2002, 14, 1391–1403. [Google Scholar] [CrossRef] [PubMed]
- Casaretto, J.; Ho, T.H. The transcription factors HvABI5 and HvVP1 are required for the abscisic acid induction of gene expression in barley aleurone cells. Plant Cell 2003, 15, 271–284. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.A.; Jungil, C.; Han, M.; Chulhyun, A.; Jongseong, J.; An, G.; Park, P.B. The ABRE-binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice. J. Plant Physiol. 2010, 167, 1512–1520. [Google Scholar] [CrossRef] [PubMed]
- Jakoby, M.; Weisshaar, B.; Drögelaser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Weltmeier, F.; Rahmani, F.; Ehlert, A.; Dietrich, K.; Schütze, K.; Wang, X.; Chaban, C.; Hanson, J.; Teige, M.; Harter, K. Expression patterns within the Arabidopsis C/S1 bZIP transcription factor network: Availability of heterodimerization partners controls gene expression during stress response and development. Plant Mol. Biol. 2009, 69, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Chattopadhyay, M.K.; Chatterjee, P.; Ghosh, B.; Sengupta, D.N. Expression of abscisic acid-responsive element-binding protein in salt-tolerant Indica rice (Oryza sativa L. cv. Pokkali). Plant Mol. Biol. 1998, 37, 629–637. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, F.; Maeta, E.; Terashima, A.; Takumi, S. Positive role of a wheat HvABI5 ortholog in abiotic stress response of seedlings. Physiol. Plant 2008, 134, 74–86. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.H.; Li, C.W.; Su, R.C.; Cheng, C.P.; Sanjaya; Tsai, Y.C.; Chan, M.T. A tomato bZIP transcription factor, SlAREB, is involved in water deficit and salt stress response. Planta 2010, 231, 1459–1473. [Google Scholar] [CrossRef] [PubMed]
- Yáñez, M.; Cáceres, S.; Orellana, S.; Bastías, A.; Verdugo, I.; Ruizlara, S.; Casaretto, J.A. An abiotic stress-responsive bZIP transcription factor from wild and cultivated tomatoes regulates stress-related genes. Plant Cell Rep. 2009, 28, 1497–1507. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Zhang, J.S.; Chen, S.Y.; Zhang, W.K. Role of Soybean GmbZIP132 under Abscisic Acid and Salt Stresses. J. Integr. Plant Biol. 2008, 50, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Choi, H.W.; Hwang, I.S.; Du, S.C.; Hwang, B.K. Functional roles of the pepper pathogen-induced bZIP transcription factor, CAbZIP1, in enhanced resistance to pathogen infection and environmental stresses. Planta 2006, 224, 1209–1225. [Google Scholar] [CrossRef] [PubMed]
- Chuang, C.F.; Running, M.P.; Williams, R.W.; Meyerowitz, E.M. The PERIANTHIA gene encodes a bZIP protein involved in the determination of floral organ number in Arabidopsis thaliana. Gene Dev. 1999, 13, 334–344. [Google Scholar] [CrossRef] [PubMed]
- Fukazawa, J.; Sakai, T.; Ishida, S.; Yamaguchi, I.; Kamiya, Y.; Takahashi, Y. Repression of shoot growth, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell 2000, 12, 901–915. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Zhu, Q.; Dai, S.; Lamb, C.; Beachy, R.N. RF2a, a bZIP transcriptional activator of the phloem-specific rice tungro bacilliform virus promoter, functions in vascular development. Embo J. 1997, 16, 5247–5259. [Google Scholar] [CrossRef] [PubMed]
- Baena-González, E.; Rolland, F.; Thevelein, J.M.; Sheen, J. A central integrator of transcription networks in plant stress and energy signalling. Nature 2007, 448, 938–942. [Google Scholar] [CrossRef] [PubMed]
- Ciceri, P.; Locatelli, F.; Genga, A.; Viotti, A.; Schmidt, R.J. The Activity of the Maize Opaque2 Transcriptional Activator Is Regulated Diurnally. Plant Physiol. 1999, 121, 1321–1328. [Google Scholar] [CrossRef] [PubMed]
- Weltmeier, F.; Ehlert, A.; Mayer, C.S.; Dietrich, K.; Wang, X.; Schütze, K.; Alonso, R.; Harter, K.; Vicente-Carbajosa, J.; Dröge-Laser, W. Combinatorial control of Arabidopsis proline dehydrogenase transcription by specific heterodimerisation of bZIP transcription factors. Embo J. 2006, 25, 3133–3143. [Google Scholar] [CrossRef] [PubMed]
- Iwata, Y.; Koizumi, N. An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc. Natl. Acad. Sci. USA 2005, 102, 5280–5285. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.X.; Srivastava, R.; Che, P.; Howell, S.H. Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J. 2007, 51, 897–909. [Google Scholar] [CrossRef] [PubMed]
- Lara, P.; Oñate-Sánchez, L.; Abraham, Z.; Ferrándiz, C.; Díaz, I.; Carbonero, P.; Vicente-Carbajosa, J. Synergistic Activation of Seed Storage Protein Gene Expression in Arabidopsis by ABI3 and Two bZIPs Related to OPAQUE2. J. Biol. Chem. 2003, 278, 21003–21011. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.C.; Ren, H.B.; He, X.; Ma, Z.Y.; Fan, C. Identification and characterization of bZIP-type transcription factors involved in carrot (Daucus carota L.) somatic embryogenesis. Plant J. 2009, 60, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef] [PubMed]
- Jenczewski, E.; Chèvre, A.; Alix, K. Chromosomal and gene expression changes in Brassica Allopolyploids. In Polyploid and Hybrid Genomics; Chen, J., Birchle, J., Eds.; John Wiley and Sons, Inc.: Oxford, IA, USA, 2013; pp. 171–186. [Google Scholar] [CrossRef]
- Lee, T.H.; Tang, H.; Wang, X.; Paterson, A.H. PGDD: A database of gene and genome duplication in plants. Nucleic Acids Res. 2013, 41, D1152–D1158. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhou, J.; Zhang, B.; Vanitha, J.; Ramachandran, S.; Jiang, S.Y. Genome-wide Expansion and Expression Divergence of the Basic Leucine Zipper Transcription Factors in Higher Plants with an Emphasis on Sorghum. J. Plant Ecol. 2011, 53, 212–231. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, N.; Chen, F.; Cai, B.; Santo, S.D.; Tornielli, G.B.; Pezzotti, M.; Cheng, Z.M. Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis vinifera). BMC Genom. 2014, 15, 281. [Google Scholar] [CrossRef] [PubMed]
- Baloglu, M.C.; Eldem, V.; Hajyzadeh, M.; Unver, T. Genome-wide analysis of the bZIP transcription factors in cucumber. PLoS ONE 2014, 9, e96014. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Xu, W.; Liu, A. Genomic surveys and expression analysis of bZIP gene family in castor bean (Ricinus communis L.). Planta 2014, 239, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Pourabed, E.; Ghane, G.F.; Soleymani, M.P.; Razavi, S.M.; Shobbar, Z.S. Basic Leucine Zipper Family in Barley: Genome-Wide Characterization of Members and Expression Analysis. Mol. Biotechnol. 2015, 57, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Qu, C.; Fu, F.; Liu, M.; Zhao, H.; Liu, C.; Li, J.; Tang, Z.; Xu, X.; Qiu, X.; Wang, R. Comparative Transcriptome Analysis of Recessive Male Sterility (RGMS) in Sterile and Fertile Brassica napus Lines. PLoS ONE 2015, 10, e0144118. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Wang, Y.; Liu, Z.; Cheng, H.; Xue, Y. HemI: A Toolkit for Illustrating Heatmaps. PLoS ONE 2014, 9, e111988. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Zhang, L.; Wu, Y.H.; Cao, Y.L.; Lu, C.M. Comparison of Five Endogenous Reference Genes for Specific PCR Detection and Quantification of Brassica napus. J. Agric. Food Chem. 2010, 58, 2812–2817. [Google Scholar] [CrossRef] [PubMed]
- Sankoff, D.; Zheng, C.; Zhu, Q. The collapse of gene complement following whole genome duplication. BMC Genom. 2010, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lydiate, D.J.; Parkin, I.A.; Falentin, C.; Delourme, R.; Carion, P.W.; King, G.J. Integration of linkage maps for the Amphidiploid Brassica napus and comparative mapping with Arabidopsis and Brassica rapa. BMC Genom. 2011, 12, 101. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chu, Z. Genome-wide evolutionary characterization and analysis of bZIP transcription factors and their expression profiles in response to multiple abiotic stresses in Brachypodium distachyon. BMC Genom. 2015, 16, 227. [Google Scholar] [CrossRef] [PubMed]
- Peviani, A.; Lastdrager, J.; Hanson, J.; Snel, B. The phylogeny of C/S1 bZIP transcription factors reveals a shared algal ancestry and the pre-angiosperm translational regulation of S1 transcripts. Sci. Rep. 2016, 6, 30444. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Yang, H.; Yan, Y.; Wei, Y.; Tie, W.; Ding, Z.; Jiao, Z.; Peng, M.; Li, K. Genome-wide characterization and analysis of bZIP transcription factor gene family related to abiotic stress in cassava. Sci. Rep. 2016, 6, 22783. [Google Scholar] [CrossRef] [PubMed]
- Lan, T.; Gao, J.; Zeng, Q.Y. Genome-wide analysis of the LEA (late embryogenesis abundant) protein gene family in Populus trichocarpa. Tree Genet. Genomes 2013, 9, 253–264. [Google Scholar] [CrossRef]
- Liang, Y.; Xiong, Z.; Zheng, J.; Xu, D.; Zhu, Z.; Xiang, J.; Gan, J.; Raboanatahiry, N.; Yin, Y.; Li, M. Genome-wide identification, structural analysis and new insights into late embryogenesis abundant (LEA) gene family formation pattern in Brassica napus. Sci. Rep. 2016, 6, 24265. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Guo, R.; Guo, C.; Hou, H.; Wang, X.; Gao, H. Evolutionary and Expression Analyses of the Apple Basic Leucine Zipper Transcription Factor Family. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Nuruzzaman, M.; Manimekalai, R.; Sharoni, A.M.; Satoh, K.; Kondoh, H.; Ooka, H.; Kikuchi, S. Genome-wide analysis of NAC transcription factor family in rice. Gene 2010, 465, 30–44. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Zhang, H.; Guo, C.; Cheng, C.; Guo, R.; Mao, L.; Fei, Z.; Wang, X. Evolutionary and Expression Analyses of Basic Zipper Transcription Factors in the Highly Homozygous Model Grape PN40024 (Vitis vinifera L.). Plant Mol. Biol. Rep. 2014, 32, 1085–1102. [Google Scholar] [CrossRef]
- Liang, Y.; Wan, N.; Cheng, Z.; Mo, Y.; Liu, B.; Liu, H.; Raboanatahiry, N.; Yin, Y.; Li, M. Whole-Genome Identification and Expression Pattern of the Vicinal Oxygen Chelate Family in Rapeseed (Brassica napus L.). Front. Plant Sci. 2017, 8, 745. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.Q.; Jian, H.J.; Yang, B.; Lu, K.; Zhang, A.X.; Liu, P.; Li, J.N. Genome-wide analysis and expression profiling of the GRF gene family in oilseed rape (Brassica napus L.). Gene 2017, 620, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Raboanatahiry, N.H.; Yin, Y.; Li, C.; Li, M. Genome-wide identification and Phylogenic analysis of kelch motif containing ACBP in Brassica napus. BMC Genom. 2015, 16, 512. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wang, C.; Yang, B.; Wu, F.; Hao, X.; Liang, W.; Niu, F.; Yan, J.; Zhang, H.; Wang, B. Identification and functional analysis of mitogen-activated protein kinase kinase kinase (MAPKKK) genes in canola (Brassica napus L.). J. Exp. Bot. 2014, 65, 2171–2188. [Google Scholar] [CrossRef] [PubMed]
- Yoo, M.J.; Ma, T.; Zhu, N.; Liu, L.; Harmon, A.C.; Wang, Q.; Chen, S. Genome-wide identification and homeolog-specific expression analysis of the SnRK2 genes in Brassica napus guard cells. Plant Mol. Biol. 2016, 91, 211–227. [Google Scholar] [CrossRef] [PubMed]
- Cavell, A.C.; Lydiate, D.J.; Parkin, I.A.; Dean, C.; Trick, M. Collinearity between a 30-centimorgan segment of Arabidopsis thaliana chromosome 4 and duplicated regions within the Brassica napus genome. Genome 1998, 41, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Wu, J.; Wang, X. Genome triplication drove the diversification of Brassica plants. Hortic. Res. 2014, 1, 14024. [Google Scholar] [CrossRef] [PubMed]
- Rana, D.; Boogaart, T.; O’neill, C.M.; Hynes, L.; Bent, E.; Macpherson, L.; Park, J.Y.; Lim, Y.P.; Bancroft, I. Conservation of the microstructure of genome segments in Brassica napus and its diploid relatives. Plant J. 2004, 40, 725–733. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Tehrim, S.; Zhang, F.; Tong, C.; Huang, J.; Cheng, X.; Dong, C.; Zhou, Y.V.; Qin, R.; Hua, W.; et al. Genome-wide comparative analysis of NBS-encoding genes between Brassica species and Arabidopsis thaliana. BMC Genom. 2014, 15, 3. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 2014, 5, 170. [Google Scholar] [CrossRef] [PubMed]
- Li, L.M.; Lü, S.Y.; Li, R.J. The Arabidopsis endoplasmic reticulum associated degradation pathways are involved in the regulation of heat stress response. Biochem. Biophys. Res. Commun. 2017, 487, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Restovic, F.; Espinoza-Corral, R.; Gómez, I.; Vicente-Carbajosa, J.; Jordana, X. An active Mitochondrial Complex II Present in Mature Seeds Contains an Embryo-Specific Iron—Sulfur Subunit Regulated by ABA and bZIP53 and Is Involved in Germination and Seedling Establishment. Front. Plant Sci. 2017, 8, 277. [Google Scholar] [CrossRef] [PubMed]
- Uno, Y.; Furihata, T.; Abe, H.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 2000, 97, 11632–11637. [Google Scholar] [CrossRef] [PubMed]
- Martínez-García, J.F.; Moyano, E.; Alcocer, M.J.; Martin, C. Two bZIP proteins from Antirrhinum flowers preferentially bind a hybrid C-box/G-box motif and help to define a new sub-family of bZIP transcription factors. Plant J. 1998, 13, 489–505. [Google Scholar] [CrossRef] [PubMed]
- Seong, E.S.; Kwon, S.U.; Ghimire, B.K.; Yu, C.Y.; Cho, D.H.; Lim, J.D.; Kim, K.S.; Heo, K.; Lim, E.S.; Chung, I.M. LebZIP2 induced by salt and drought stress and transient overexpression by Agrobacterium. BMB Rep. 2008, 41, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Strathmann, A.; Kuhlmann, M.; Heinekamp, T.; Drögelaser, W. BZI-1 specifically heterodimerises with the tobacco bZIP transcription factors BZI-2, BZI-3/TBZF and BZI-4, and is functionally involved in flower development. Plant J. 2001, 28, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Weiste, C.; Pedrotti, L.; Selvanayagam, J.; Muralidhara, P.; Fröschel, C.; Novák, O.; Ljung, K.; Hanson, J.; Dröge-Laser, W. The Arabidopsis bZIP11 transcription factor links low-energy signalling to auxin-mediated control of primary root growth. PLoS Genet. 2017, 13, e1006607. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Becker, D.F. Connecting proline metabolism and signaling pathways in plant senescence. Front. Plant Sci. 2015, 6, 552. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Li, F.; Dong, W.; Liu, W.; Qian, Y.; Yang, J.; Wang, F.; Wu, Y. Nicotiana benthamiana NbbZIP28, a possible regulator of unfolded protein response, plays a negative role in viral infection. Eur. J. Plant Pathol. 2017, 1–13. [Google Scholar] [CrossRef]
- Angelos, E.; Ruberti, C.; Kim, S.J.; Brandizzi, F. Maintaining the factory: The roles of the unfolded protein response in cellular homeostasis in plants. Plant J. 2017, 90, 671–682. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.X.; Srivastava, R.; Che, P.; Howell, S.H. An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell 2007, 19, 4111. [Google Scholar] [CrossRef] [PubMed]
- Popova, O.V.; Yang, O.; Dietz, K.-J.; Golldack, D. Differential transcript regulation in Arabidopsis thaliana and the halotolerant Lobularia maritima indicates genes with potential function in plant salt adaptation. Gene 2008, 423, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Yang, O.; Popova, O.V.; Süthoff, U.; Lüking, I.; Dietz, K.J.; Golldack, D. The Arabidopsis basic leucine zipper transcription factor AtbZIP24 regulates complex transcriptional networks involved in abiotic stress resistance. Gene 2009, 436, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Assunção, A.; Herrero, E.; Lin, Y.-F.; Huettel, B.; Talukdar, S.; Smaczniak, C.; Immink, R.; van Eldik, M.; Fiers, M.; Schat, H.; et al. Arabidopsis thaliana transcription factors bZIP19 and bZIP23 regulate the adaptation to zinc deficiency. Proc. Natl. Acad. Sci. USA 2010, 107, 10296–10301. [Google Scholar] [CrossRef] [PubMed]
- Castro, P.H.; Lilay, G.H.; Muñoz-Mérida, A.; Schjoerring, J.K.; Azevedo, H.; Assunção, A.G.L. Phylogenetic analysis of F-bZIP transcription factors indicates conservation of the zinc deficiency response across land plants. Sci. Rep. 2017, 7, 3806. [Google Scholar] [CrossRef] [PubMed]








© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Xu, D.; Jia, L.; Huang, X.; Ma, G.; Wang, S.; Zhu, M.; Zhang, A.; Guan, M.; Lu, K.; et al. Genome-Wide Identification and Structural Analysis of bZIP Transcription Factor Genes in Brassica napus. Genes 2017, 8, 288. https://0-doi-org.brum.beds.ac.uk/10.3390/genes8100288
Zhou Y, Xu D, Jia L, Huang X, Ma G, Wang S, Zhu M, Zhang A, Guan M, Lu K, et al. Genome-Wide Identification and Structural Analysis of bZIP Transcription Factor Genes in Brassica napus. Genes. 2017; 8(10):288. https://0-doi-org.brum.beds.ac.uk/10.3390/genes8100288
Chicago/Turabian StyleZhou, Yan, Daixiang Xu, Ledong Jia, Xiaohu Huang, Guoqiang Ma, Shuxian Wang, Meichen Zhu, Aoxiang Zhang, Mingwei Guan, Kun Lu, and et al. 2017. "Genome-Wide Identification and Structural Analysis of bZIP Transcription Factor Genes in Brassica napus" Genes 8, no. 10: 288. https://0-doi-org.brum.beds.ac.uk/10.3390/genes8100288





