A Review of Current and Emerging Approaches for Water Pollution Monitoring
Abstract
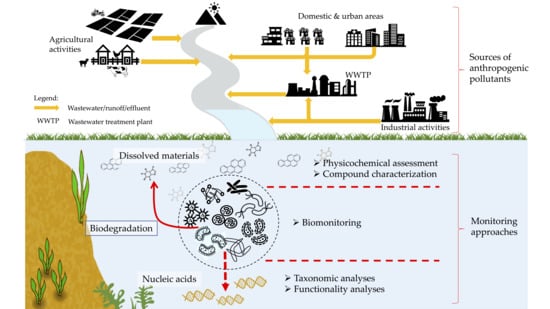
:1. Introduction
2. Utilizable Elements for Wastewater Monitoring
2.1. Physicochemical Characterization in Monitoring Environmental Pollution
2.2. Spectrometry Technology in Detecting Toxicant in Wastewaters
2.3. Biomonitoring Environmental Pollution
Reliability of Bacterial Community in Waterways Biomonitoring
2.4. Molecular Approaches in Taxonomical Analyses
2.4.1. Assessment of Microbial Community Using Molecular Fingerprinting Techniques
2.4.2. Advanced High-Throughput Sequencing for Complex Taxonomic Diversity Assessment
2.5. Gene Functionality Assessment in Aquatic Environment
2.5.1. Quantification of Functional Genes Involved in the Biodegradation of Pollutants Using qRT-PCR
2.5.2. Microarray as Environmental Monitoring Tool
2.5.3. Advanced Gene Expression Profiling for Wastewater and Aquatic System Monitoring Using Metatranscriptomics Approach
2.6. Functionality Analysis of Bacterial Nucleic Acids through Flow Cytometry
3. Summary and Future Outlooks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tilley, E.; Ulrich, L.; Lüthi, C.; Reymond, P.; Zurbrügg, C. Compendium of Sanitation Systems and Technologies, 2nd ed.; Eawag: Duebendorf, Switzerland, 2014; p. 175. [Google Scholar]
- Nuria, C.-F.; Josep, C.; Munjanja, B.; Nollet, L.; Lambropoulou, D. High-resolution mass spectrometric techniques for structural characterization and determination of organic pollutants in the environment. In Chromatographic Analysis of the Environment: Mass Spectrometry Based Approaches, 4th ed.; Nollet, L.M., Lambropoulou, D.A., Eds.; CRC Press: Boca Raton, FL, USA, 2017; Volume 3, pp. 47–78. [Google Scholar]
- Şener, Ş.; Şener, E.; Davraz, A. Evaluation of water quality using water quality index (WQI) method and GIS in Aksu River (SW-Turkey). Sci. Total Environ. 2017, 584, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Stachler, E.; Akyon, B.; De Carvalho, N.A.; Ference, C.; Bibby, K. Correlation of crAssphage qPCR markers with culturable and molecular indicators of human fecal pollution in an impacted urban watershed. Environ. Sci. Technol. 2018, 52, 7505–7512. [Google Scholar] [CrossRef] [PubMed]
- Bondaruk, J.; Janson, E.; Wysocka, M.; Chalupnik, S. Identification of hazards for water environment in the upper Silesian Coal Basin caused by the discharge of salt mine water containing particularly harmful substances and radionuclides. J. Sustain. Min. 2015, 14, 179–187. [Google Scholar] [CrossRef] [Green Version]
- Harry, I.S.K.; Ameh, E.; Coulon, F.; Nocker, A. Impact of treated sewage effluent on the microbiology of a small brook using flow cytometry as a diagnostic tool. Water Air Soil Pollut. 2016, 227, 57. [Google Scholar] [CrossRef] [PubMed]
- WWAP (United Nations World Water Assessment Programme). Executive summary. In The United Nations World Water Development Report 2017: Wastewater: The Untapped Resource; Connor, R., Renata, A., Ortigara, C., Koncagül, E., Uhlenbrook, S., Lamizana-Diallo, B.M., Zadeh, S.M., Qadir, M., Kjellén, M., Sjödin, J., Eds.; UNESCO: Paris, France, 2017; p. 2. [Google Scholar]
- Hollender, J.; Van Bavel, B.; Dulio, V.; Farmen, E.; Furtmann, K.; Koschorreck, J.; Kunkel, U.; Krauss, M.; Munthe, J.; Schlabach, M.; et al. High-resolution mass spectrometry-based non-target screening can support regulatory environmental monitoring and chemicals management. Environ. Sci. Eur. 2019, 31, 42. [Google Scholar] [CrossRef] [Green Version]
- Carstea, E.M.; Bridgeman, J.; Baker, A.; Reynolds, D.M. Fluorescence spectroscopy for wastewater monitoring: A review. Water Res. 2016, 95, 205–219. [Google Scholar] [CrossRef]
- Diamantini, E.; Lutz, S.R.; Mallucci, S.; Majone, B.; Merz, R.; Bellin, A. Driver detection of water quality trends in three large European river basins. Sci. Total Environ. 2018, 612, 49–62. [Google Scholar] [CrossRef]
- Meng, F.; Huang, G.; Yang, X.; Li, Z.; Li, J.; Cao, J.; Wang, Z.; Sun, L. Identifying the sources and fate of anthropogenically impacted dissolved organic matter (DOM) in urbanized rivers. Water Res. 2013, 47, 5027–5039. [Google Scholar] [CrossRef]
- Michael-Kordatou, I.; Michael, C.; Duan, X.; He, X.; Dionysiou, D.; Mills, M.; Fatta-Kassinos, D. Dissolved effluent organic matter: Characteristics and potential implications in wastewater treatment and reuse applications. Water Res. 2015, 77, 213–248. [Google Scholar] [CrossRef]
- Bolyard, S.C.; Reinhart, D. Evaluation of leachate dissolved organic nitrogen discharge effect on wastewater effluent quality. Waste Manag. 2017, 65, 47–53. [Google Scholar] [CrossRef]
- Mesfioui, R.; Love, N.G.; Bronk, D.A.; Mulholland, M.R.; Hatcher, P.G. Reactivity and chemical characterization of effluent organic nitrogen from wastewater treatment plants determined by Fourier transform ion cyclotron resonance mass spectrometry. Water Res. 2012, 46, 622–634. [Google Scholar] [CrossRef] [PubMed]
- Maizel, A.C.; Remucal, C.K. The effect of advanced secondary municipal wastewater treatment on the molecular composition of dissolved organic matter. Water Res. 2017, 122, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Guo, L. Intriguing changes in molecular size and composition of dissolved organic matter induced by microbial degradation and self-assembly. Water Res. 2018, 135, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Patel, N.; Khan, M.D.; Shahane, S.; Rai, D.; Chauhan, D.; Kant, C.; Chaudhary, V.K. Emerging pollutants in aquatic environment: Source, effect, and challenges in biomonitoring and bioremediation—A review. Pollution 2020, 6, 99–113. [Google Scholar] [CrossRef]
- Tiwari, B.; Drogui, P.; Tyagi, R. Removal of emerging micro-pollutants from pharmaceutical industry wastewater. In Current Developments in Biotechnology and Bioengineering; Elsevier BV: Amsterdam, The Netherlands, 2020; pp. 457–480. [Google Scholar]
- Department of Environment (DOE). Available online: https://www.doe.gov.my/portalv1/wp-content/uploads/2019/05/Standard-Kualiti-Air-Kebangsaan.pdf (accessed on 28 January 2020).
- Horton, R.K. An index number system for rating water quality. J. Water Pollut. Control Fed. 1965, 37, 300–306. [Google Scholar]
- Tyagi, S.; Sharma, B.; Singh, P.; Dobhal, R. Water quality assessment in terms of water quality index. Am. J. Water Resour. 2020, 1, 34–38. [Google Scholar] [CrossRef]
- Srivastava, G.; Kumar, P. Water quality index with missing parameters. Int. J. Res. Eng. Technol. 2013, 2, 609–614. [Google Scholar]
- Abbasnia, A.; Yousefi, N.; Mahvi, A.H.; Nabizadeh, R.; Radfard, M.; Yousefi, M.; Alimohammadi, M. Evaluation of groundwater quality using water quality index and its suitability for assessing water for drinking and irrigation purposes: Case study of Sistan and Baluchistan province (Iran). Hum. Ecol. Risk Assess. Int. J. 2018, 25, 988–1005. [Google Scholar] [CrossRef]
- Meher, P.K.; Sharma, P.; Gautam, Y.P.; Kumar, A.; Mishra, K.P. Evaluation of water quality of Ganges River using water quality index tool. Environ. Asia 2015, 8, 124–132. [Google Scholar]
- Brown, R.M.; McClelland, N.I.; Deininger, R.A.; Tozer, R.G. A water quality index- do we dare? Water Sewage Work. 1970, 117, 339–343. [Google Scholar]
- Saffran, K.; Cash, K.; Hallard, K.; Neary, B.; Wright, C. CCME water quality index 1.0 user’s manual. Canadian Water Quality Guidelines for The Protection of Aquatic Life, Canadian Environmental Quality Guidelines; Canadian Council of Ministers of the Environment: Winnipeg, MB, Canada, 2001. [Google Scholar]
- Cude, C.G. Oregon water quality index a tool for evaluating water quality management effectiveness. JAWRA J. Am. Water Resour. Assoc. 2001, 37, 125–137. [Google Scholar] [CrossRef]
- Al-Shujairi, S.O.H. Develop and apply water quality index to evaluate water quality of Tigris and Euphrates Rivers in Iraq. Int. J. Mod. Eng. Res. 2013, 3, 2119–2126. [Google Scholar]
- Laurenson, G.; Laurenson, S.; Bolan, N.; Beecham, S.; Clark, I. The role of bioretention systems in the treatment of stormwater. In Advances in Agronomy; Elsevier BV: Amsterdam, The Netherlands, 2013; Volume 120, pp. 223–274. [Google Scholar]
- Wu, J.; Ren, Y.; Wang, X.; Wang, X.; Chen, L.; Liu, G. Nitrogen and phosphorus associating with different size suspended solids in roof and road runoff in Beijing, China. Environ. Sci. Pollut. Res. 2015, 22, 15788–15795. [Google Scholar] [CrossRef] [PubMed]
- Ballo, S.; Liu, M.; Hou, L.; Chang, J. Pollutants in stormwater runoff in Shanghai (China): Implications for management of urban runoff pollution. Prog. Nat. Sci. 2009, 19, 873–880. [Google Scholar] [CrossRef]
- Pillai, H.; Girish, K. Rubber processing industry effluent treatment using a bacterial consortium. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 775–782. [Google Scholar]
- Elsergany, M.; Ahsan, A.; Aziz, M.A. Optimizing the performance of a paper mill effluent treatment. Sains Malays. 2015, 44, 101–106. [Google Scholar] [CrossRef]
- Phalakornkule, C.; Polgumhang, S.; Tongdaung, W.; Karakat, B.; Nuyut, T. Electrocoagulation of blue reactive, red disperse and mixed dyes, and application in treating textile effluent. J. Environ. Manag. 2010, 91, 918–926. [Google Scholar] [CrossRef] [PubMed]
- Bello, M.; Nourouzi, M.; Abdullah, L.C.; Choong, T.S.; Koay, Y.; Keshani, S. POME is treated for removal of color from biologically treated POME in fixed bed column: Applying wavelet neural network (WNN). J. Hazard. Mater. 2013, 262, 106–113. [Google Scholar] [CrossRef]
- Napoleão, D.; Zaidan, L.M.C.; Rodríguez-Díaz, J.; da Rocha Santana, R.; de Mendonça, M.B.d.S.; da Nova Araújo, A.; Benachour, M.; da Silva, V.L. Use of the photo-Fenton process to discover the degradation of drugs present in water from the wastewater treatment plants of the pharmaceutical industry. Afinidad 2018, 75, 23–31. [Google Scholar]
- Ramdani, N.; Benouis, K.; Lousdad, A.; Hamou, A.; Boufadi, M.Y. Physicochemical and bacteriological characterization of hospital effluents and their impact on the environment. Chem. Int. 2018, 4, 102–108. [Google Scholar]
- Noutsopoulos, C.; Andreadakis, A.; Kouris, N.; Charchousi, D.; Mendrinou, P.; Galani, A.; Mantziaras, I.; Koumaki, E. Greywater characterization and loadings—Physicochemical treatment to promote onsite reuse. J. Environ. Manag. 2018, 216, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Liu, J.; Aini, G.; Gong, Y. A comparative study of the grain-size distribution of surface dust and stormwater runoff quality on typical urban roads and roofs in Beijing, China. Environ. Sci. Pollut. Res. 2016, 23, 2693–2704. [Google Scholar] [CrossRef] [PubMed]
- De Mello, K.; Valente, R.A.; Randhir, T.O.; Vettorazzi, C.A. Impacts of tropical forest cover on water quality in agricultural watersheds in Southeastern Brazil. Ecol. Indic. 2018, 93, 1293–1301. [Google Scholar] [CrossRef]
- Dikonketso, S.M.; Rasheed, A.; Olayinka, A.A. The analysis of physicochemical characteristics of pig farm seepage and its possible impact on the receiving natural environment. Afr. J. Environ. Sci. Technol. 2016, 10, 242–252. [Google Scholar] [CrossRef] [Green Version]
- Omofunmi, O.; Ilesanmi, O.; Alli, A. Assessing catfish pond effluent on soil physicochemical properties and its suitability for crop production. Arid Zone J. Eng. Technol. Environ. 2018, 14, 355–366. [Google Scholar]
- Huang, X.-F.; Ye, G.-Y.; Yi, N.-K.; Lu, L.-J.; Zhang, L.; Yang, L.-Y.; Xiao, L.; Liu, J. Effect of plant physiological characteristics on the removal of conventional and emerging pollutants from aquaculture wastewater by constructed wetlands. Ecol. Eng. 2019, 135, 45–53. [Google Scholar] [CrossRef]
- Chang, Y.-T.; Chang, Y.-J. The investigation of the bacterial indicators and point sources of pollution for the Nanshih River, Taiwan: A case study. Desalin. Water Treat. 2013, 52, 1130–1142. [Google Scholar] [CrossRef]
- Nazeer, S.; Hashmi, M.Z.; Malik, R.N. Heavy metals distribution, risk assessment and water quality characterization by water quality index of the River Soan, Pakistan. Ecol. Indic. 2014, 43, 262–270. [Google Scholar] [CrossRef]
- Yang, L.; Han, D.H.; Lee, B.-M.; Hur, J. Characterizing treated wastewaters of different industries using clustered fluorescence EEM–PARAFAC and FT-IR spectroscopy: Implications for downstream impact and source identification. Chemosphere 2015, 127, 222–228. [Google Scholar] [CrossRef]
- Van Loosdrecht, M.C.M.; Brdjanovic, D. Anticipating the next century of wastewater treatment. Science 2014, 344, 1452–1453. [Google Scholar] [CrossRef]
- Ruff, M.; Mueller, M.S.; Loos, M.; Singer, H. Quantitative target and systematic non-target analysis of polar organic micro-pollutants along the river Rhine using high-resolution mass-spectrometry—Identification of unknown sources and compounds. Water Res. 2015, 87, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Bade, R.; Bijlsma, L.; Sancho, J.V.; Baz-Lomba, J.A.; Castiglioni, S.; Castrignanò, E.; Causanilles, A.; Gracia-Lor, E.; Kasprzyk-Hordern, B.; Kinyua, J.; et al. Liquid chromatography-tandem mass spectrometry determination of synthetic cathinones and phenethylamines in influent wastewater of eight European cities. Chemosphere 2017, 168, 1032–1041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamjunke, N.; Von Tümpling, W.; Hertkorn, N.; Harir, M.; Schmitt-Kopplin, P.; Norf, H.; Weitere, M.; Herzsprung, P. A new approach for evaluating transformations of dissolved organic matter (DOM) via high-resolution mass spectrometry and relating it to bacterial activity. Water Res. 2017, 123, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; He, C.; Li, Y.; Chung, K.H.; Xu, C.; Shi, Q. Fractionation and characterization of dissolved organic matter (DOM) in refinery wastewater by revised phase retention and ion-exchange adsorption solid phase extraction followed by ESI FT-ICR MS. Talanta 2017, 162, 466–473. [Google Scholar] [CrossRef] [PubMed]
- Verkh, Y.; Rožman, M.; Petrovic, M. A non-targeted high-resolution mass spectrometry data analysis of dissolved organic matter in wastewater treatment. Chemosphere 2018, 200, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Mohd-Nor, D.; Ramli, N.; Sharuddin, S.S.; Hassan, M.A.; Mustapha, N.A.; Ariffin, H.; Sakai, K.; Tashiro, Y.; Shirai, Y.; Maeda, T. Dynamics of microbial populations responsible for biodegradation during the full-scale treatment of palm oil mill effluent. Microbes Environ. 2019, 34, 121–128. [Google Scholar] [CrossRef] [Green Version]
- Eggen, R.I.L.; Hollender, J.; Joss, A.; Schärer, M.; Stamm, C. Reducing the discharge of micropollutants in the aquatic environment: The benefits of upgrading wastewater treatment plants. Environ. Sci. Technol. 2014, 48, 7683–7689. [Google Scholar] [CrossRef]
- Brack, W.; Hollender, J.; De Alda, M.L.; Müller, C.; Schulze, T.; Schymanski, E.; Slobodnik, J.; Krauss, M. High-resolution mass spectrometry to complement monitoring and track emerging chemicals and pollution trends in European water resources. Environ. Sci. Eur. 2019, 31, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Hernández, E.H.; Andrades, M.; Álvarez-Martín, A.; Pose-Juan, E.; Rodríguez-Cruz, M.; Sánchez-Martín, M. Occurrence of pesticides and some of their degradation products in waters in a Spanish wine region. J. Hydrol. 2013, 486, 234–245. [Google Scholar] [CrossRef]
- Duong, H.T.; Kadokami, K.; Pan, S.; Matsuura, N.; Nguyen, T.Q. Screening and analysis of 940 organic micro-pollutants in river sediments in Vietnam using an automated identification and quantification database system for GC–MS. Chemosphere 2014, 107, 462–472. [Google Scholar] [CrossRef]
- Robles-Molina, J.; Gilbert-López, B.; García-Reyes, J.F.; Molina-Díaz, A. Monitoring of selected priority and emerging contaminants in the Guadalquivir River and other related surface waters in the province of Jaén, South East Spain. Sci. Total Environ. 2014, 479, 247–257. [Google Scholar] [CrossRef]
- Kong, L.; Kadokami, K.; Wang, S.; Duong, H.T.; Chau, H.T.C. Monitoring of 1300 organic micro-pollutants in surface waters from Tianjin, North China. Chemosphere 2015, 122, 125–130. [Google Scholar] [CrossRef]
- Hernández, F.; Sancho, J.V.; Ibáñez, M.; Abad, E.; Portolés, T.; Mattioli, L. Current use of high-resolution mass spectrometry in the environmental sciences. Anal. Bioanal. Chem. 2012, 403, 1251–1264. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Wang, D.; Tang, Y.; Xing, X.; Zhuang, J.; Cheng, J.; Du, Z. Fabric-phase sorptive extraction coupled with ion mobility spectrometry for on-site rapid detection of PAHs in aquatic environment. Talanta 2019, 195, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Siddig, A.A.; Ellison, A.M.; Ochs, A.; Villar-Leeman, C.; Lau, M.K. How do ecologists select and use indicator species to monitor ecological change? Insights from 14 years of publication in Ecological Indicators. Ecol. Indic. 2016, 60, 223–230. [Google Scholar] [CrossRef] [Green Version]
- Holt, E.A.; Miller, S.W. Bioindicators: Using organisms to measure environmental impacts. Nat. Educ. Knowl. 2011, 3, 8. [Google Scholar]
- Mccarty, L.; Power, M.; Munkittrick, K.R. Bioindicators versus biomarkers in ecological risk assessment. Hum. Ecol. Risk Assess. Int. J. 2002, 8, 159–164. [Google Scholar] [CrossRef]
- Adams, S.M.; Giesy, J.P.; Tremblay, L.A.; Eason, C. The use of biomarkers in ecological risk assessment: Recommendations from the Christchurch conference on biomarkers in ecotoxicology. Biomarkers 2001, 6, 1–6. [Google Scholar] [CrossRef]
- Marzin, A.; Archaimbault, V.; Belliard, J.; Chauvin, C.; Delmas, F.; Pont, D. Ecological assessment of running waters: Do macrophytes, macroinvertebrates, diatoms and fish show similar responses to human pressures? Ecol. Indic. 2012, 23, 56–65. [Google Scholar] [CrossRef]
- Lainé, M.; Morin, S.; Tison-Rosebery, J. A multicompartment approach - diatoms, macrophytes, benthic macroinvertebrates and fish - to assess the impact of toxic industrial releases on a small French river. PLoS ONE 2014, 9, e102358. [Google Scholar] [CrossRef]
- Green, H.C.; Haugland, R.A.; Varma, M.; Millen, H.T.; Borchardt, M.A.; Field, K.G.; Walters, W.A.; Knight, R.; Sivaganesan, M.; Kelty, C.A.; et al. Improved HF183 quantitative real-time PCR assay for characterization of human fecal pollution in ambient surface water samples. Appl. Environ. Microbiol. 2014, 80, 3086–3094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, W.; Sidhu, J.P.S.; Smith, K.; Beale, D.J.; Gyawali, P.; Toze, S. Distributions of fecal markers in wastewater from different climatic zones for human fecal pollution tracking in Australian surface waters. Appl. Environ. Microbiol. 2015, 82, 1316–1323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rousk, J.; Bååth, E.; Brookes, P.C.; Lauber, C.L.; Lozupone, C.; Caporaso, J.G.; Knight, R.; Fierer, N. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010, 4, 1340–1351. [Google Scholar] [CrossRef] [PubMed]
- Bates, S.T.; Berg-Lyons, D.; Caporaso, J.G.; Walters, W.; Knight, R.; Fierer, N. Examining the global distribution of dominant archaeal populations in soil. ISME J. 2011, 5, 908–917. [Google Scholar] [CrossRef] [Green Version]
- Fierer, N.; Leff, J.W.; Adams, B.J.; Nielsen, U.N.; Bates, S.T.; Lauber, C.L.; Owens, S.; Gilbert, J.A.; Wall, D.H.; Caporaso, J.G. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. USA 2012, 109, 21390–21395. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Zhao, L.; Lu, W.; Jia, H.; Sun, Y. Bacterial communities in water and sediment shaped by paper mill pollution and indicated bacterial taxa in sediment in Daling River. Ecol. Indic. 2016, 60, 766–773. [Google Scholar] [CrossRef]
- Lee, D.-Y.; Lee, H.; Trevors, J.T.; Weir, S.C.; Thomas, J.L.; Habash, M. Characterization of sources and loadings of fecal pollutants using microbial source tracking assays in urban and rural areas of the Grand River Watershed, Southwestern Ontario. Water Res. 2014, 53, 123–131. [Google Scholar] [CrossRef]
- Wood, S.A.; Smith, K.F.; Banks, J.C.; Tremblay, L.A.; Rhodes, L.L.; Mountfort, D.O.; Cary, S.C.; Pochon, X. Molecular genetic tools for environmental monitoring of New Zealand’s aquatic habitats, past, present and the future. N. Z. J. Mar. Freshw. Res. 2013, 47, 90–119. [Google Scholar] [CrossRef] [Green Version]
- Priac, A.; Badot, P.-M.; Crini, G. Treated wastewater phytotoxicity assessment using Lactuca sativa: Focus on germination and root elongation test parameters. Comptes Rendus Biol. 2017, 340, 188–194. [Google Scholar] [CrossRef]
- Charles, J.; Sancey, B.; Morin-Crini, N.; Badot, P.-M.; Degiorgi, F.; Trunfio, G.; Crini, G. Evaluation of the phytotoxicity of polycontaminated industrial effluents using the lettuce plant (Lactuca sativa) as a bioindicator. Ecotoxicol. Environ. Saf. 2011, 74, 2057–2064. [Google Scholar] [CrossRef]
- Ladislas, S.; El-Mufleh, A.; Gérente, C.; Chazarenc, F.; Andres, Y.; Béchet, B. Potential of aquatic macrophytes as bioindicators of heavy metal pollution in urban stormwater runoff. Water Air Soil Pollut. 2011, 223, 877–888. [Google Scholar] [CrossRef]
- Farias, D.; Hurd, C.; Eriksen, R.; MacLeod, C. Macrophytes as bioindicators of heavy metal pollution in estuarine and coastal environments. Mar. Pollut. Bull. 2018, 128, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Klink, A.; Polechońska, L.; Cegłowska, A.; Stankiewicz, A. Typha latifolia (broadleaf cattail) as bioindicator of different types of pollution in aquatic ecosystems—Application of self-organizing feature map (neural network). Environ. Sci. Pollut. Res. 2016, 23, 14078–14086. [Google Scholar] [CrossRef] [PubMed]
- Baeta, A.; Vieira, L.R.; Lírio, A.V.; Canhoto, C.; Marques, J.C.; Guilhermino, L. Use of stable isotope ratios of fish larvae as indicators to assess diets and patterns of anthropogenic nitrogen pollution in estuarine ecosystems. Ecol. Indic. 2017, 83, 112–121. [Google Scholar] [CrossRef] [Green Version]
- Omar, W.A.; Mikhail, W.; Abdo, H.M.; El Defan, T.A.A.; Poraas, M.M. Ecological risk assessment of metal pollution along Greater Cairo Sector of the River Nile, Egypt, using nile tilapia, Oreochromis niloticus, as bioindicator. J. Toxicol. 2015, 2015, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.-C.; Lu, P.-N.; Huang, H.-L.; Chu, C.; Li, H.-P.; Tsai, H.-J. Zebrafish transgenic line huORFZ is an effective living bioindicator for detecting environmental toxicants. PLoS ONE 2014, 9, e90160. [Google Scholar] [CrossRef] [Green Version]
- Kumar, N.; Krishnani, K.K.; Singh, N.P. Oxidative and cellular stress as bioindicators for metal contamination in freshwater mollusk Lamellidens marginalis. Environ. Sci. Pollut. Res. 2017, 24, 16137–16147. [Google Scholar] [CrossRef]
- Júnior, C.D.S.M.; Juen, L.; Hamada, N. Analysis of urban impacts on aquatic habitats in the central Amazon basin: Adult odonates as bioindicators of environmental quality. Ecol. Indic. 2015, 48, 303–311. [Google Scholar] [CrossRef]
- Bråte, I.L.N.; Hurley, R.; Iversen, K.; Beyer, J.; Thomas, K.V.; Steindal, C.C.; Green, N.W.; Olsen, M.; Lusher, A. Mytilus spp. as sentinels for monitoring microplastic pollution in Norwegian coastal waters: A qualitative and quantitative study. Environ. Pollut. 2018, 243, 383–393. [Google Scholar] [CrossRef]
- Mahaut, M.-L.; Basuyaux, O.; Baudinière, E.; Chataignier, C.; Pain, J.; Caplat, C. The porifera Hymeniacidon perlevis (Montagu, 1818) as a bioindicator for water quality monitoring. Environ. Sci. Pollut. Res. 2012, 20, 2984–2992. [Google Scholar] [CrossRef]
- Wu, H.; Chen, P.; Tsay, T. Assessment of nematode community structure as a bioindicator in river monitoring. Environ. Pollut. 2010, 158, 1741–1747. [Google Scholar] [CrossRef] [PubMed]
- Filimonova, V.; Gonçalves, F.J.M.; Marques, J.C.; De Troch, M.; Gonçalves, F.J.M. Biochemical and toxicological effects of organic (herbicide Primextra® Gold TZ) and inorganic (copper) compounds on zooplankton and phytoplankton species. Aquat. Toxicol. 2016, 177, 33–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Zhang, M. Primary consumers as bioindicator of nitrogen pollution in lake planktonic and benthic food webs. Ecol. Indic. 2012, 14, 189–196. [Google Scholar] [CrossRef]
- Neves, M.F.J.V.; Castro, B.B.; Vidal, T.; Vieira, R.H.S.D.F.; Marques, J.C.; Coutinho, J.A.P.; Goncalves, F.; Gonçalves, A. Biochemical and populational responses of an aquatic bioindicator species, Daphnia longispina, to a commercial formulation of a herbicide (Primextra® Gold TZ) and its active ingredient (S-metolachlor). Ecol. Indic. 2015, 53, 220–230. [Google Scholar] [CrossRef]
- Hamza, I.A.; Jurzik, L.; Überla, K.; Wilhelm, M. Evaluation of pepper mild mottle virus, human picobirnavirus and Torque teno virus as indicators of fecal contamination in river water. Water Res. 2011, 45, 1358–1368. [Google Scholar] [CrossRef]
- Yau, V.M.; Schiff, K.C.; Arnold, B.F.; Griffith, J.F.; Gruber, J.S.; Wright, C.C.; Wade, T.J.; Burns, S.; Hayes, J.M.; McGee, C.; et al. Effect of submarine groundwater discharge on bacterial indicators and swimmer health at Avalon Beach, CA, USA. Water Res. 2014, 59, 23–36. [Google Scholar] [CrossRef]
- Zhang, Y.; Kelly, W.R.; Panno, S.V.; Liu, Y.-Q. Tracing fecal pollution sources in karst groundwater by Bacteroidales genetic biomarkers, bacterial indicators, and environmental variables. Sci. Total Environ. 2014, 490, 1082–1090. [Google Scholar] [CrossRef]
- Sharuddin, S.S.; Ramli, N.; Hassan, M.A.; Mustapha, N.A.; Amran, A.; Mohd-Nor, D.; Sakai, K.; Tashiro, Y.; Shirai, Y.; Maeda, T. Bacterial community shift revealed Chromatiaceae and Alcaligenaceae as potential bioindicators in the receiving river due to palm oil mill effluent final discharge. Ecol. Indic. 2017, 82, 526–529. [Google Scholar] [CrossRef]
- Acosta, M.D.L.R.; Jiménez-Collazo, J.; Maldonado-Román, M.; Malavé-Llamas, K.; Musa-Wasil, J.C. Bacteria as potential indicators of heavy metal contamination in a tropical mangrove and the implications on environmental and human health. J. Trop. Life Sci. 2015, 5, 110–116. [Google Scholar] [CrossRef]
- Zamyadi, A.; McQuaid, N.; Prévost, M.; Dorner, S. Monitoring of potentially toxic cyanobacteria using an online multi-probe in drinking water sources. J. Environ. Monit. 2012, 14, 579–588. [Google Scholar] [CrossRef]
- Artigas, J.; Arts, G.H.; Babut, M.; Caracciolo, A.B.; Charles, S.; Chaumot, A.; Combourieu, B.; Dahllöf, I.; Despréaux, D.; Ferrari, B.J.D.; et al. Towards a renewed research agenda in ecotoxicology. Environ. Pollut. 2012, 160, 201–206. [Google Scholar] [CrossRef]
- Tlili, A.; Berard, A.; Blanck, H.; Bouchez, A.; Cássio, F.; Eriksson, K.M.; Morin, S.; Montuelle, B.; Navarro, E.; Pascoal, C.; et al. Pollution-induced community tolerance (PICT): Towards an ecologically relevant risk assessment of chemicals in aquatic systems. Freshw. Biol. 2015, 61, 2141–2151. [Google Scholar] [CrossRef] [Green Version]
- Clements, W.H.; Rohr, J.R. Community responses to contaminants: Using basic ecological principles to predict ecotoxicological effects. Environ. Toxicol. Chem. 2009, 28, 1789–1800. [Google Scholar] [CrossRef] [PubMed]
- Wakelin, S.A.; Gerard, E.; Van Koten, C.; Banabas, M.; O’Callaghan, M.; Nelson, P.N. Soil physicochemical properties impact more strongly on bacteria and fungi than conversion of grassland to oil palm. Pedobiology 2016, 59, 83–91. [Google Scholar] [CrossRef]
- Bolaji, E.J.; Opeyemi, I.E.; Eniola, O.O. Impact of palm oil mill effluent on physico-chemical parameters of a Southwestern River, Ekiti State, Nigeria. In 2nd Africa Regional Conference Technical Proceedings; Derkyi, M., Awuah, E., Obeng-Ofori, D., Derkyi, N.S.A., Owusu-Ansah, F., Eds.; University of Energy and Natural Resources: Sunyani, Ghana, 2015; pp. 638–646. [Google Scholar]
- Gözdereliler, E.; Boon, N.; Aamand, J.; De Roy, K.; Granitsiotis, M.S.; Albrechtsen, H.-J.; Sørensen, S.R. Comparing metabolic functionalities, community structures, and dynamics of herbicide-degrading communities cultivated with different substrate concentrations. Appl. Environ. Microbiol. 2012, 79, 367–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, L.; Zhang, T. Bacterial communities in different sections of a municipal wastewater treatment plant revealed by 16S rDNA 454 pyrosequencing. Appl. Microbiol. Biotechnol. 2013, 97, 2681–2690. [Google Scholar] [CrossRef] [Green Version]
- Nshimyimana, J.P.; Ekklesia, E.; Shanahan, P.; Chua, L.H.C.; Thompson, J.R. Distribution and abundance of human-specific Bacteroides and relation to traditional indicators in an urban tropical catchment. J. Appl. Microbiol. 2014, 116, 1369–1383. [Google Scholar] [CrossRef] [Green Version]
- Ryall, B.; Eydallin, G.; Ferenci, T. Culture history and population heterogeneity as determinants of bacterial adaptation: The adaptomics of a single environmental transition. Microbiol. Mol. Biol. Rev. 2012, 76, 597–625. [Google Scholar] [CrossRef] [Green Version]
- Mlejnková, H.; Sovová, K. Impact of pollution and seasonal changes on microbial community structure in surface water. Water Sci. Technol. 2010, 61, 2787–2795. [Google Scholar] [CrossRef]
- Zhao, J.; Zhao, X.; Chao, L.; Zhang, W.; You, T.; Zhang, J. Diversity change of microbial communities responding to zinc and arsenic pollution in a river of northeastern China. J. Zhejiang Univ. Sci. B 2014, 15, 670–680. [Google Scholar] [CrossRef] [Green Version]
- Lear, G.; Ancion, P.-Y.; Harding, J.; Lewis, G. Use of bacterial communities to assess the ecological health of a recently restored stream. N. Z. J. Mar. Freshw. Res. 2012, 46, 291–301. [Google Scholar] [CrossRef]
- Witt, V.; Wild, C.; Uthicke, S. Effect of substrate type on bacterial community composition in biofilms from the Great Barrier Reef. FEMS Microbiol. Lett. 2011, 323, 188–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kriwy, P.; Uthicke, S. Microbial diversity in marine biofilms along a water quality gradient on the Great Barrier Reef. Syst. Appl. Microbiol. 2011, 34, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Borrero-Santiago, A.R.; Bautista-Chamizo, E.; DelValls, T.; Riba, I. A possible CO2 leakage event: Can the marine microbial community be recovered? Mar. Pollut. Bull. 2017, 117, 380–385. [Google Scholar] [CrossRef]
- Niu, L.; Li, Y.; Wang, P.; Zhang, W.; Wang, C.; Li, J.; Wu, H. Development of a microbial community-based index of biotic integrity (MC-IBI) for the assessment of ecological status of rivers in the Taihu Basin, China. Ecol. Indic. 2018, 85, 204–213. [Google Scholar] [CrossRef]
- Paulino, G.V.B.; Félix, C.R.; Silvan, C.G.; Andersen, G.L.; Landell, M.F. Bacterial community and environmental factors associated to rivers runoff and their possible impacts on coral reef conservation. Mar. Pollut. Bull. 2020, 156, 111233. [Google Scholar] [CrossRef] [PubMed]
- Shahi, A.; Ince, B.; Aydin, S.; Ince, O. Assessment of the horizontal transfer of functional genes as a suitable approach for evaluation of the bioremediation potential of petroleum-contaminated sites: A mini-review. Appl. Microbiol. Biotechnol. 2017, 101, 4341–4348. [Google Scholar] [CrossRef]
- Stewart, E.J. Growing Unculturable Bacteria. J. Bacteriol. 2012, 194, 4151–4160. [Google Scholar] [CrossRef] [Green Version]
- De Tender, C.; Schlundt, C.; Devriese, L.I.; Mincer, T.J.; Zettler, E.R.; Amaral-Zettler, L. A review of microscopy and comparative molecular-based methods to characterize “Plastisphere” communities. Anal. Methods 2017, 9, 2132–2143. [Google Scholar] [CrossRef]
- Lear, G.; Dopheide, A.; Ancion, P.; Lewis, G.D. A comparison of bacterial, ciliate and macroinvertebrate indicators of stream ecological health. Aquat. Ecol. 2011, 45, 517–527. [Google Scholar] [CrossRef]
- Díez, B.; Pedrós-Alió, C.; Marsh, T.L.; Massana, R. Application of Denaturing Gradient Gel Electrophoresis (DGGE) To Study the Diversity of Marine Picoeukaryotic Assemblages and Comparison of DGGE with Other Molecular Techniques. Appl. Environ. Microbiol. 2001, 67, 2942–2951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duarte, S.; Cássio, F.; Pascoal, C. Denaturing gradient gel electrophoresis (DGGE) in Microbial Ecology—Insights from Freshwaters. In Gel Electrophoresis—Principles and Basics; IntechOpen: London, UK, 2012; pp. 173–196. [Google Scholar]
- Nielsen, K.L.; Godfrey, P.A.; Stegger, M.; Andersen, P.S.; Feldgarden, M.; Frimodt-Møller, N. Selection of unique Escherichia coli clones by random amplified polymorphic DNA (RAPD): Evaluation by whole genome sequencing. J. Microbiol. Methods 2014, 103, 101–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sjöberg, F.; Nowrouzian, F.; Rangel, I.; Hannoun, C.; Moore, E.; Adlerberth, I.; Wold, A.E. Comparison between terminal-restriction fragment length polymorphism (T-RFLP) and quantitative culture for analysis of infants’ gut microbiota. J. Microbiol. Methods 2013, 94, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Moreno, L.I.; Mills, D.; Fetscher, J.; John-Williams, K.; Meadows-Jantz, L.; Mccord, B. The application of amplicon length heterogeneity PCR (LH-PCR) for monitoring the dynamics of soil microbial communities associated with cadaver decomposition. J. Microbiol. Methods 2011, 84, 388–393. [Google Scholar] [CrossRef] [PubMed]
- Naknim, V.; Kutanan, W.; Lomthaisong, K. Identifying the origin of forensic soil evidence using amplified ribosomal DNA restriction analysis of its bacterial community. Chiang Mai Univ. J. Nat. Sci. 2016, 15, 115–128. [Google Scholar] [CrossRef]
- Ciesielski, S.; Bułkowska, K.; Dabrowska, D.; Kaczmarczyk, D.; Kowal, P.; Możejko-Ciesielska, J. Ribosomal intergenic spacer analysis as a tool for monitoring methanogenic archaea changes in an anaerobic digester. Curr. Microbiol. 2013, 67, 240–248. [Google Scholar] [CrossRef] [Green Version]
- Gao, G.; Yin, D.; Chen, S.; Xia, F.; Yang, J.; Li, Q.; Wang, W. Effect of biocontrol agent Pseudomonas fluorescens 2P24 on soil fungal community in cucumber rhizosphere using T-RFLP and DGGE. PLoS ONE 2012, 7, e31806. [Google Scholar] [CrossRef]
- Zhang, W.; Mo, Y.; Yang, J.; Zhou, J.; Lin, Y.; Isabwe, A.; Zhang, J.; Gao, X.; Yu, Z. Genetic diversity pattern of microeukaryotic communities and its relationship with the environment based on PCR-DGGE and T-RFLP techniques in Dongshan Bay, Southeast China. Cont. Shelf Res. 2018, 164, 1–9. [Google Scholar] [CrossRef]
- Vallaeys, T.; Topp, E.; Muyzer, G.; Macheret, V.; Laguerre, G.; Rigaud, A.; Soulas, G. Evaluation of denaturing gradient gel electrophoresis in the detection of 16S rDNA sequence variation in rhizobia and methanotrophs. FEMS Microbiol. Ecol. 2006, 24, 279–285. [Google Scholar] [CrossRef]
- Muyzer, G.; Smalla, K. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie van Leeuwenhoek 1998, 73, 127–141. [Google Scholar] [CrossRef]
- Cetecioglu, Z.; Ince, O.; Ince, B. Gel Electrophoresis Based Genetic Fingerprinting Techniques on Environmental Ecology. In Gel Electrophoresis—Advanced Techniques; IntechOpen: London, UK, 2012; pp. 52–66. [Google Scholar]
- Du, H.; Harata, N.; Li, F. Responses of riverbed sediment bacteria to heavy metals: Integrated evaluation based on bacterial density, activity and community structure under well-controlled sequencing batch incubation conditions. Water Res. 2018, 130, 115–126. [Google Scholar] [CrossRef]
- Chonova, T.; Labanowski, J.; Cournoyer, B.; Chardon, C.; Keck, F.; Laurent, É.; Mondamert, L.; Vasselon, V.; Wiest, L.; Bouchez, A. River biofilm community changes related to pharmaceutical loads emitted by a wastewater treatment plant. Environ. Sci. Pollut. Res. 2017, 25, 9254–9264. [Google Scholar] [CrossRef]
- Smalla, K.; Oros-Sichler, M.; Milling, A.; Heuer, H.; Baumgarte, S.; Becker, R.; Neuber, G.; Kropf, S.; Ulrich, A.; Tebbe, C.C. Bacterial diversity of soils assessed by DGGE, T-RFLP and SSCP fingerprints of PCR-amplified 16S rRNA gene fragments: Do the different methods provide similar results? J. Microbiol. Methods 2007, 69, 470–479. [Google Scholar] [CrossRef]
- Van Elsas, J.D.; Boersma, F.G.H. A review of molecular methods to study the microbiota of soil and the mycosphere. Eur. J. Soil Biol. 2011, 47, 77–87. [Google Scholar] [CrossRef]
- Karczewski, K.; Riss, H.W.; Meyer, E.I. Comparison of DNA-fingerprinting (T-RFLP) and high-throughput sequencing (HTS) to assess the diversity and composition of microbial communities in groundwater ecosystems. Limnologica 2017, 67, 45–53. [Google Scholar] [CrossRef]
- Ramette, A.N. Quantitative community fingerprinting methods for estimating the abundance of operational taxonomic units in natural microbial communities. Appl. Environ. Microbiol. 2009, 75, 2495–2505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindström, S.; Rowe, O.; Timonen, S.; Sundström, L.; Johansson, H. Trends in bacterial and fungal communities in ant nests observed with terminal-restriction fragment length polymorphism (T-RFLP) and Next Generation Sequencing (NGS) techniques—Validity and compatibility in ecological studies. PeerJ 2018, 6, e5289. [Google Scholar] [CrossRef]
- Loman, N.J.; Constantinidou, C.; Chan, J.Z.M.; Halachev, M.R.; Sergeant, M.J.; Penn, C.W.; Robinson, E.R.; Pallen, M.J. High-throughput bacterial genome sequencing: An embarrassment of choice, a world of opportunity. Nat. Rev. Genet. 2012, 10, 599–606. [Google Scholar] [CrossRef]
- Deviram, G.; Charankumar, B.; Ramu, K.; Madeswaran, P.; Murthy, M.V.R. A review on the applications and recent advances in environmental DNA (eDNA) metagenomics. Rev. Environ. Sci. Bio Technol. 2019, 18, 389–411. [Google Scholar] [CrossRef]
- Glenn, T.C. Field guide to next-generation DNA sequencers. Mol. Ecol. Resour. 2011, 11, 759–769. [Google Scholar] [CrossRef]
- Jünemann, S.; Sedlazeck, F.J.; Prior, K.; Albersmeier, A.; John, U.; Kalinowski, J.; Mellmann, A.; Goesmann, A.; Von Haeseler, A.; Stoye, J.; et al. Updating benchtop sequencing performance comparison. Nat. Biotechnol. 2013, 31, 294–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mardis, E.R. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008, 24, 133–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mason, O.U.; Scott, N.M.; Gonzalez, A.P.; Robbins-Pianka, A.; Baelum, J.; Kimbrel, J.A.; Bouskill, N.J.; Prestat, E.; Borglin, S.E.; Joyner, D.C.; et al. Metagenomics reveals sediment microbial community response to Deepwater Horizon oil spill. ISME J. 2014, 8, 1464–1475. [Google Scholar] [CrossRef] [Green Version]
- Mohd-Nor, D.; Ramli, N.; Sharuddin, S.S.; Hassan, M.A.; Mustapha, N.A.; Amran, A.; Sakai, K.; Shirai, Y.; Maeda, T. Alcaligenaceae and Chromatiaceae as reliable bioindicators present in palm oil mill effluent final discharge treated by different biotreatment processes. Ecol. Indic. 2018, 95, 468–473. [Google Scholar] [CrossRef]
- Albertsen, M.; Hansen, L.B.S.; Saunders, A.M.; Nielsen, P.H.; Nielsen, K.L. A metagenome of a full-scale microbial community carrying out enhanced biological phosphorus removal. ISME J. 2012, 6, 1094–1106. [Google Scholar] [CrossRef]
- Ju, F.; Guo, F.; Ye, L.; Xia, Y.; Zhang, T. Metagenomic analysis on seasonal microbial variations of activated sludge from a full-scale wastewater treatment plant over 4 years. Environ. Microbiol. Rep. 2014, 6, 80–89. [Google Scholar] [CrossRef]
- Guo, J.; Peng, Y.; Ni, B.-J.; Han, X.; Fan, L.; Yuan, Z. Dissecting microbial community structure and methane-producing pathways of a full-scale anaerobic reactor digesting activated sludge from wastewater treatment by metagenomic sequencing. Microb. Cell Factories 2015, 14, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Wang, C.; Zhang, W.; Di, P.; Yi, N.; Chen, C. Vertical and horizontal assemblage patterns of bacterial communities in a eutrophic river receiving domestic wastewater in southeast China. Environ. Pollut. 2017, 230, 469–478. [Google Scholar] [CrossRef]
- Ouyang, L.; Chen, H.; Liu, X.; Wong, M.H.; Xu, F.; Yang, X.; Xu, W.; Zeng, Q.; Wang, W.; Li, S. Characteristics of spatial and seasonal bacterial community structures in a river under anthropogenic disturbances. Environ. Pollut. 2020, 264, 114818. [Google Scholar] [CrossRef]
- Zhang, K.; Zhang, Z.; Hu, Z.; Zeng, F.; Chen, C.; Yang, X.; Li, Y. Bacterial community composition and function succession under aerobic and anaerobic conditions impacts the biodegradation of 17β-estradiol and its environmental risk. Environ. Pollut. 2020, 267, 115155. [Google Scholar] [CrossRef]
- Jin, D.; Kong, X.; Cui, B.; Jin, S.; Xie, Y.; Wang, X.; Deng, Y. Bacterial communities and potential waterborne pathogens within the typical urban surface waters. Sci. Rep. 2018, 8, 13368. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.C.; Messina, M.A.; Nicolosi, G.; Petralia, S.; Baker, M.D.; Mayne, C.K.S.; Dinon, C.M.; Moss, C.J.; Onac, B.P.; Garey, J.R.; et al. Surface runoff alters cave microbial community structure and function. PLoS ONE 2020, 15, e0232742. [Google Scholar] [CrossRef] [PubMed]
- Zolkefli, N.; Ramli, N.; Mohamad-Zainal, N.S.L.; Mustapha, N.A.; Yusoff, M.Z.M.; Hassan, M.A.; Maeda, T. Alcaligenaceae and Chromatiaceae as pollution bacterial bioindicators in palm oil mill effluent (POME) final discharge polluted rivers. Ecol. Indic. 2020, 111, 106048. [Google Scholar] [CrossRef]
- Cordier, T.; Lanzén, A.; Apothéloz-Perret-Gentil, L.; Stoeck, T.; Pawlowski, J. Embracing environmental genomics and machine learning for routine biomonitoring. Trends Microbiol. 2019, 27, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Jaspers, E.; Overmann, J. Ecological significance of microdiversity: Identical 16S rRNA gene sequences can be found in bacteria with highly divergent genomes and ecophysiologies. Appl. Environ. Microbiol. 2004, 70, 4831–4839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basile, L.A.; Erijman, L. Quantitative assessment of phenol hydroxylase diversity in bioreactors using a functional gene analysis. Appl. Microbiol. Biotechnol. 2008, 78, 863–872. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Perry, N.; Chuahan, A.; Sayler, G.; Ripp, S. Microbial indicators for monitoring pollution and bioremediation. In Microbial Biodegradation and Bioremediation; Elsevier BV: Amsterdam, The Netherlands, 2014; pp. 115–136. [Google Scholar]
- Hasenbein, M.; Werner, I.; Deanovic, L.A.; Geist, J.; Fritsch, E.B.; Javidmehr, A.; Foe, C.; Fangue, N.A.; Connon, R.E. Transcriptomic profiling permits the identification of pollutant sources and effects in ambient water samples. Sci. Total Environ. 2014, 468, 688–698. [Google Scholar] [CrossRef]
- Rani, B.; Sharma, V. Transcriptome profiling: Methods and applications—A review. Agric. Rev. 2017, 38, 271–281. [Google Scholar] [CrossRef] [Green Version]
- Juskowiak, B. Nucleic acid-based fluorescent probes and their analytical potential. Anal. Bioanal. Chem. 2011, 399, 3157–3176. [Google Scholar] [CrossRef] [Green Version]
- De Boer, P.; Rahaoui, H.; Leer, R.; Montijn, R.; Van Der Vossen, J. Real-time PCR detection of Campylobacter spp.: A comparison to classic culturing and enrichment. Food Microbiol. 2015, 51, 96–100. [Google Scholar] [CrossRef]
- González-Plaza, J.J.; Blau, K.; Milaković, M.; Jurina, T.; Smalla, K.; Udikovic-Kolic, N. Antibiotic-manufacturing sites are hot-spots for the release and spread of antibiotic resistance genes and mobile genetic elements in receiving aquatic environments. Environ. Int. 2019, 130, 104735. [Google Scholar] [CrossRef] [PubMed]
- Tolosi, R.; Apostolakos, I.; Laconi, A.; Carraro, L.; Grilli, G.; Cagnardi, P.; Piccirillo, A. Rapid detection and quantification of plasmid-mediated colistin resistance genes (mcr-1 to mcr-5) by real-time PCR in bacterial and environmental samples. J. Appl. Microbiol. 2020, 129, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Hafeez, F.; Martin-Laurent, F.; Béguet, J.; Bru, D.; Cortet, J.; Schwartz, C.; Morel, J.-L.; Philippot, L. Taxonomic and functional characterization of microbial communities in technosols constructed for remediation of a contaminated industrial wasteland. J. Soils Sediments 2012, 12, 1396–1406. [Google Scholar] [CrossRef]
- Keshri, J.; Yousuf, B.; Mishra, A.; Jha, B. The abundance of functional genes, cbbL, nifH, amoA and apsA, and bacterial community structure of intertidal soil from Arabian Sea. Microbiol. Res. 2015, 175, 57–66. [Google Scholar] [CrossRef]
- Zhao, H.-M.; Hu, R.-W.; Du, H.; Xin, X.-P.; Li, Y.-W.; Li, H.; Cai, Q.-Y.; Mo, C.-H.; Liu, J.-S.; Zhou, D.; et al. Functional genomic analysis of phthalate acid ester (PAE) catabolism genes in the versatile PAE-mineralising bacterium Rhodococcus sp. 2G. Sci. Total Environ. 2018, 640, 646–652. [Google Scholar] [CrossRef]
- Chakraborty, A.; Dasgupta, C.K.; Bhadury, P. Application of molecular techniques for the assessment of microbial communities in contaminated sites. In Microbial Biodegradation and Bioremediation; Elsevier BV: Amsterdam, The Netherlands, 2014; pp. 85–113. [Google Scholar]
- Stefano, G.B.; Mantione, K.; Kream, R.M.; Kuzelova, H.; Ptáček, R.; Raboch, J.; Samuel, J.M. Comparing bioinformatic gene expression profiling methods: Microarray and RNA-Seq. Med. Sci. Monit. Basic Res. 2014, 20, 138–142. [Google Scholar] [CrossRef] [Green Version]
- He, Z.; Deng, Y.; Zhou, J. Development of functional gene microarrays for microbial community analysis. Curr. Opin. Biotechnol. 2012, 23, 49–55. [Google Scholar] [CrossRef]
- Jie, S.; Li, M.; Gan, M.; Zhu, J.; Yin, H.; Liu, X. Microbial functional genes enriched in the Xiangjiang River sediments with heavy metal contamination. BMC Microbiol. 2016, 16, 179. [Google Scholar] [CrossRef] [Green Version]
- Cong, J.; Yang, Y.; Liu, X.; Lu, H.; Liu, X.; Zhou, J.; Li, D.; Yin, H.; Ding, J.; Zhang, Y. Analyses of soil microbial community compositions and functional genes reveal potential consequences of natural forest succession. Sci. Rep. 2015, 5, 10007. [Google Scholar] [CrossRef] [Green Version]
- Short, M.D.; Abell, G.C.; Bodrossy, L.; Akker, B.V.D. Application of a novel functional gene microarray to probe the functional ecology of ammonia oxidation in nitrifying activated sludge. PLoS ONE 2013, 8, e77139. [Google Scholar] [CrossRef] [Green Version]
- Chan, Y.; Van Nostrand, J.D.; Zhou, J.; Pointing, S.B.; Farrell, R.L. Functional ecology of an Antarctic Dry Valley. Proc. Natl. Acad. Sci. USA 2013, 110, 8990–8995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Nostrand, J.D.; He, Z.; Zhou, J. Use of functional gene arrays for elucidating in situ biodegradation. Front. Microbiol. 2012, 3, 339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tu, Q.; Yu, H.; He, Z.; Deng, Y.; Wu, L.; Van Nostrand, J.D.; Zhou, A.; Voordeckers, J.; Lee, Y.-J.; Qin, Y.; et al. GeoChip 4: A functional gene-array-based high-throughput environmental technology for microbial community analysis. Mol. Ecol. Resour. 2014, 14, 914–928. [Google Scholar] [CrossRef] [PubMed]
- Peimbert, M.; Alcaraz, L.D. A hitchhiker’s guide to metatranscriptomics. In Field Guidelines for Genetic Experimental Designs in High-Throughput Sequencing; Aransay, A., Lavín Trueba, J., Eds.; Springer: Cham, Switzerland, 2016; pp. 313–342. [Google Scholar]
- Croucher, N.J.; Thomson, N.R. Studying bacterial transcriptomes using RNA-seq. Curr. Opin. Microbiol. 2010, 13, 619–624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bashiardes, S.; Zilberman-Schapira, G.; Elinav, E. Use of Metatranscriptomics in microbiome research. Bioinform. Biol. Insights 2016, 10, BBI.S34610-25. [Google Scholar] [CrossRef] [Green Version]
- Crovadore, J.; Soljan, V.; Calmin, G.; Chablais, R.; Cochard, B.; Lefort, F. Metatranscriptomic and metagenomic description of the bacterial nitrogen metabolism in waste water wet oxidation effluents. Heliyon 2017, 3, e00427. [Google Scholar] [CrossRef]
- Desta, A.F.; Nzioki, J.; Maina, S.; Stomeo, F. Molecular biomonitoring of microbial communities in tannery wastewater treatment plant for the removal of retanning chemicals. Biol. Wastewater Treat. Resour. Recovery 2017, 141. [Google Scholar] [CrossRef]
- Johnson, D.R.; Lee, T.K.; Park, J.; Fenner, K.; Helbling, D.E. The functional and taxonomic richness of wastewater treatment plant microbial communities are associated with each other and with ambient nitrogen and carbon availability. Environ. Microbiol. 2014, 17, 4851–4860. [Google Scholar] [CrossRef]
- Ju, F.; Beck, K.; Yin, X.; Maccagnan, A.; McArdell, C.S.; Singer, H.P.; Johnson, D.R.; Zhang, T.; Bürgmann, H. Wastewater treatment plant resistomes are shaped by bacterial composition, genetic exchange, and upregulated expression in the effluent microbiomes. ISME J. 2019, 13, 346–360. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Klümper, U.; Liu, Y.; Yang, Y.; Wei, Q.; Lin, J.-G.; Gu, J.; Li, M. Metagenomic and metatranscriptomic analyses reveal activity and hosts of antibiotic resistance genes in activated sludge. Environ. Int. 2019, 129, 208–220. [Google Scholar] [CrossRef]
- Falk, N.; Reid, T.; Skoyles, A.; Grgicak-Mannion, A.; Drouillard, K.; Weisener, C. Microbial metatranscriptomic investigations across contaminant gradients of the Detroit River. Sci. Total Environ. 2019, 690, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Zhang, T. Metagenomic and metatranscriptomic analysis of microbial community structure and gene expression of activated sludge. PLoS ONE 2012, 7, e38183. [Google Scholar] [CrossRef] [Green Version]
- Handley, K.M.; Piceno, Y.M.; Hu, P.; Tom, L.M.; Mason, O.U.; Andersen, G.L.; Jansson, J.K.; Gilbert, J.A. Metabolic and spatio-taxonomic response of uncultivated seafloor bacteria following the Deepwater Horizon oil spill. ISME J. 2017, 11, 2569–2583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Gao, W.; Johnson, R.H.; Zhang, W.; Meldrum, D.R. Integrated metagenomic and metatranscriptomic analyses of microbial communities in the meso- and bathypelagic realm of North Pacific Ocean. Mar. Drugs 2013, 11, 3777–3801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satinsky, B.M.; Fortunato, C.S.; Doherty, M.; Smith, C.B.; Sharma, S.; Ward, N.D.; Krusche, A.V.; Yager, P.L.; Richey, J.E.; Moran, M.A.; et al. Metagenomic and metatranscriptomic inventories of the lower Amazon River, May 2011. Microbiome 2015, 3, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fortunato, C.S.; Crump, B.C. Microbial gene abundance and expression patterns across a river to ocean salinity gradient. PLoS ONE 2015, 10, e0140578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, Y.; Yamamoto, N.; Kino, Y.; Yamamoto, N.; Kamei, S.; Mori, H.; Kurokawa, K.; Nakashima, N. Establishment of a multi-species biofilm model and metatranscriptomic analysis of biofilm and planktonic cell communities. Appl. Microbiol. Biotechnol. 2016, 100, 7263–7279. [Google Scholar] [CrossRef]
- Lau, M.C.Y.; Harris, R.L.; Oh, Y.; Yi, M.J.; Behmard, A.; Onstott, T.C. Taxonomic and functional compositions impacted by the quality of metatranscriptomic assemblies. Front. Microbiol. 2018, 9, 1235. [Google Scholar] [CrossRef] [Green Version]
- Etan, B.; Ng, C.M.; Nshimyimana, J.P.; Eloh, L.-L.; Gin, K.Y.-H.; Thompson, J.R. Next-generation sequencing (NGS) for assessment of microbial water quality: Current progress, challenges, and future opportunities. Front. Microbiol. 2015, 6, 1027. [Google Scholar] [CrossRef]
- Aiken, G. Fluorescence and dissolved organic matter: A chemist’s perspective. In Aquatic Organic Matter Fluorescence; Coble, P.G., Lead, J., Baker, A., Reynolds, D.M., Spencer, R.G.M., Eds.; Cambridge University Press: New York, NY, USA, 2014; Chapter 2; pp. 35–74. [Google Scholar]
- Manti, A.; Boi, P.; Falcioni, T.; Canonico, B.; Ventura, A.; Sisti, D.; Pianetti, A.; Balsamo, M.; Papa, S. Bacterial Cell Monitoring in Wastewater Treatment Plants by Flow Cytometry. Water Environ. Res. 2008, 80, 346–354. [Google Scholar] [CrossRef]
- Hammes, F.; Egli, T. Cytometric methods for measuring bacteria in water: Advantages, pitfalls and applications. Anal. Bioanal. Chem. 2010, 397, 1083–1095. [Google Scholar] [CrossRef] [PubMed]
- Lautenschlager, K.; Hwang, C.; Liu, W.-T.; Boon, N.; Köster, O.; Vrouwenvelder, H.; Egli, T.; Hammes, F. A microbiology-based multi-parametric approach towards assessing biological stability in drinking water distribution networks. Water Res. 2013, 47, 3015–3025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gillespie, S.; Lipphaus, P.; Green, J.; Parsons, S.; Weir, P.; Juskowiak, K.; Jefferson, B.; Jarvis, P.; Nocker, A. Assessing microbiological water quality in drinking water distribution systems with disinfectant residual using flow cytometry. Water Res. 2014, 65, 224–234. [Google Scholar] [CrossRef] [PubMed]
- Nescerecka, A.; Juhna, T.; Hammes, F. Identifying the underlying causes of biological instability in a full-scale drinking water supply system. Water Res. 2018, 135, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ma, D.; Ma, L.; Song, Y.; Gao, G.; Wang, Y. Geographic distribution pattern of low and high nucleic acid content bacteria on a river-catchment scale. Mar. Freshw. Res. 2017, 68, 1618. [Google Scholar] [CrossRef] [Green Version]
- Sharuddin, S.S.; Ramli, N.; Mohd-Nor, D.; Hassan, M.A.; Maeda, T.; Shirai, Y.; Sakai, K.; Tashiro, Y. Shift of low to high nucleic acid bacteria as a potential bioindicator for the screening of anthropogenic effects in a receiving river due to palm oil mill effluent final discharge. Ecol. Indic. 2018, 85, 79–84. [Google Scholar] [CrossRef]
- Santos, M.; Oliveira, H.; Pereira, J.L.; Pereira, M.J.; Goncalves, F.; Vidal, T. Flow cytometry analysis of low/high DNA content (LNA/HNA) bacteria as bioindicator of water quality evaluation. Ecol. Indic. 2019, 103, 774–781. [Google Scholar] [CrossRef]
- Abzazou, T.; Salvadó, H.; Bruguera-Casamada, C.; Simón, P.; Lardín, C.; Araujo, R.M. Assessment of total bacterial cells in extended aeration activated sludge plants using flow cytometry as a microbial monitoring tool. Environ. Sci. Pollut. Res. 2015, 22, 11446–11455. [Google Scholar] [CrossRef]
- Foladori, P.; Bruni, L.; Tamburini, S.; Ziglio, G. Direct quantification of bacterial biomass in influent, effluent and activated sludge of wastewater treatment plants by using flow cytometry. Water Res. 2010, 44, 3807–3818. [Google Scholar] [CrossRef]
- Kim, J.M.; Lee, H.J.; Kim, S.Y.; Song, J.J.; Park, W.; Jeon, C.O. Analysis of the fine-scale population structure of “Candidatus Accumulibacter phosphatis” in enhanced biological phosphorus removal sludge, using fluorescence in situ hybridization and flow cytometric sorting. Appl. Environ. Microbiol. 2010, 76, 3825–3835. [Google Scholar] [CrossRef] [Green Version]
- Vital, M.; Stucki, D.; Egli, T.; Hammes, F. Evaluating the growth potential of pathogenic bacteria in water. Appl. Environ. Microbiol. 2010, 76, 6477–6484. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| NSFWQI [25] | CCME WQI [26] | OWQI [27] | MMWQI [19] | ||||
|---|---|---|---|---|---|---|---|
| 91–100 | Excellent | 95–100 | Excellent | 90–100 | Excellent | 90–100 | Excellent |
| 71–90 | Good | 80–94 | Good | 85–89 | Good | 80–89 | Good |
| 51–70 | Medium | 60–79 | Fair | 80–84 | Fair | 50–79 | Moderate |
| 26–50 | Bad | 45–59 | Marginal | 60–79 | Poor | 0–49 | Poor |
| 0–25 | Very bad | 0–44 | Poor | 0–59 | Very poor | ||
| (a) Industrial Wastewaters | |||||||
| * Parameters | Rubber Effluent [32] | Paper Mill Effluent [33] | Textile Effluent [34] | Raw Palm Oil Mill Effluent; POME [35] | Pharmaceutical Industry Wastewater [36] | ||
| pH | 5.7 ± 0.3 | 5.6–5.8 | 8.3–9.5 | 3.4–5.2 | 9.79 | ||
| BOD | 1340 ± 2.0 | 5279 | 137 | 10,250–43,750 | 586.07 | ||
| COD | 2834 ± 1.9 | 9507 | 278–736 | 15,000–100,000 | 1458.03 | ||
| TSS | 3512 ± 4.8 | 1747 | 85–354 | 5000–54,000 | - | ||
| TS | - | - | - | - | 407.67 | ||
| TFS | - | - | - | - | 162.67 | ||
| TVS | - | - | - | - | 384 | ||
| (b) Domestic Wastewater | |||||||
| * Parameters | Domestic Sewage [36] | Hospital Liquid Effluent [37] | Greywater [38] | ||||
| Dish Washer | Laundry | Kitchen | |||||
| pH | 7.6 | 7.45–8.85 | 10 ± 0.2 | 8.3 ± 0.8 | 6.9 ± 0.4 | ||
| EC (μS/cm) | - | 152–350 | 2199 ± 753 | 653 ± 423 | 449 ± 341 | ||
| BOD | 600 | 255–850 | 184.6 ± 24 | 1363 ± 950 | 831 ± 358 | ||
| COD | 1006.42 | 455–879 | 411 ± 59 | 2072 ± 1401 | 1119 ± 476 | ||
| TSS | - | 420–640 | 11 ± 1.3 | 169 ± 96 | 319 ± 209 | ||
| VSS | - | - | 10 ± 0.5 | 139 ± 90 | 314 ± 205 | ||
| TS | 888 | - | 2535 ± 1053 | 1085 ± 608 | 883 ± 426 | ||
| TFS | 504 | - | - | - | - | ||
| TVS | 245 | - | - | - | - | ||
| NH4 + | - | 7–47.9 | - | - | - | ||
| NH4+-N | - | - | 0.11 ± 0.07 | 1.4 ± 1.1 | 0.20 ± 0.26 | ||
| NO2– | - | 2.1–4.2 | - | - | - | ||
| NO3– | - | 23.9–56.7 | - | - | - | ||
| NO3––N | - | - | 0.44 ± 0.06 | 0.68 ± 0.53 | 1.1 ± 1.2 | ||
| NO2––N (μg/L) | - | - | 0.05 ± 0.01 | 75 ± 56 | 20 ± 17 | ||
| TN | - | - | <0.5 | 6.2 ± 5.3 | 6.5 ± 5.0 | ||
| TP | - | - | 187 ± 51 | 1.2 ± 0.81 | 2.7 ± 3.1 | ||
| PO4–P | - | - | <0.05 | 0.22 ± 0.66 | 1.5 ± 2.8 | ||
| (c) Agricultural Wastewater and Stormwater Runoffs | |||||||
| * Parameters | Agricultural Wastewaters | Stormwater Runoffs [39] | |||||
| Degraded Agricultural Watershed [40] | Swine Wastewater [41] | Catfish Pond Water [42] | Aqua-Culture Wastewater [43] | Rooftop runoff | Road Runoff in Residen-tial Area | Main Traffic Road Run-off | |
| pH | - | 6.5–9 | 6.86 ± 0.06 | - | - | - | - |
| EC (mS/cm) | - | 1.25–5.58 | 2.92 ± 1.1 | - | - | - | - |
| DO | 7.91 ± 0.61 | 4.14–7.64 | 7.50 ± 1.10 | 2.45 ± 0.13 | - | - | - |
| BOD | - | 163–3550 | 1.82 ± 0.12 | - | - | - | - |
| COD | - | 210–9400 | - | 66.6 ± 6.44 | 346.92 ± 241.88 | 561.75 ± 719.30 | 570.70 ± 489.41 |
| TSS | 21.13 ± 28.41 | - | 486.20 ± 11.60 | - | 43.07 ± 31.78 | 286.94 ± 187.96 | 373.77 ± 186.23 |
| TDS | - | 770–648 | 602.60 ± 15. 80 | - | - | - | - |
| NTU | 29.66 ± 24.5 | 0.21–3.65 | 112.5 ± 4.6 | - | - | - | - |
| ISS | 15.14 ± 23.31 | - | - | - | - | - | - |
| NH4+-N | - | - | - | 2.35 ± 0.56 | 23.88 ± 17.24 | 21.44 ± 22.48 | 15.29 ± 10.04 |
| NO3––N | - | - | - | 0.51 ± 0.013 | 19.06 ± 12.49 | 11.41 ± 10.58 | 9.94 ± 6.98 |
| NO2––N | - | - | - | 0.134 ± 0.03 | - | - | - |
| TN | 0.30 ± 0.32 | - | - | 3.60 ± 1.31 | - | - | - |
| TP | 0.09 ± 85.25 | - | - | 0.23 ± 0.047 | 0.11 ± 0.11 | 1.23 ± 1.73 | 0.16 ± 0.20 |
| PO43− | - | 55–1680 | 1.34 ± 0.05 | - | - | - | - |
| NO3 | - | 37–2730 | 2.32 ± 0.09 | - | - | - | - |
| NO2 | - | 50–1427 | - | - | - | - | - |
| Chloride | - | - | 24.75 ± 1.21 | - | - | - | - |
| Alkalinity | - | - | 101.12 ± 2.12 | - | - | - | - |
| Cu | - | - | - | - | 0.05 ± 0.06 | 0.15 ± 0.24 | 0.09 ± 0.10 |
| Fe | - | - | - | - | 0.22 ± 0.36 | 0.09 ± 0.08 | 0.11 ± 0.26 |
| Mn | - | - | - | - | 0.20 ± 0.33 | 0.16 ± 0.17 | 0.14 ± 0.12 |
| Zn | - | - | - | - | 4.40 ± 7.52 | 0.11 ± 0.05 | 0.06 ± 0.05 |
| Pb | - | - | - | - | 0.03 ± 0.03 | 0.01 ± 0.01 | 0.01 ± 0.02 |
| Bioindicator(s) | Applications | Site of Study | References | |
|---|---|---|---|---|
| Plants (Macrophytes) | Lactuca sativa | Heavy metals in industrial effluent Polycontaminated industrial effluents | Decontamination stations’ outlet of companies in Franche-Comte’, France | [76,77] |
| Oenanthe sp., Juncus sp., Typha sp., Callitriche sp. 1, Callitriche sp. 2 | Cd, Cr, Cu, Ni, Pb, Zn, and As | Storm water runoff in detention pond, northeast of Nantes, France | [78] | |
| Seagrasses: Zostera muelleri, Zostera nigricaulis, Ruppia megacarpa | As, Cd, Cu, Pb, Se, and Zn | Derwent estuary in Tasmania, Australia | [79] | |
| Typha latifolia (broadleaf cattail) | Trace metals (Cd, Cu, Fe, Mn, Ni, Pb, and Zn) from various types of pollutions | Different types of surface water resources in Greater Poland, upper and lower Silesia, Poland, Europe | [80] | |
| Macroalgae | Red: Gracillaria sp., Pyropia columbina, Porphyra lucassi, Grateloupia turuturu Brown: Scytopsiphon lomentaria, Ecklonia radiata, Undaria pinnatifida Green: Ulva australis, Ulva compressa | As, Cd, Cu, Pb, Se, and Zn | Derwent estuary in Tasmania, Australia | [79] |
| Vertebrate | Pomatoschistus microps larvae | Nitrogen contamination in estuarine ecosystem | Minho and Lima estuaries in Portugal | [81] |
| Oreochromis niloticus | Metals (Cu, Zn, Mn, Cd, Pb, and Fe) | River Nile, Egypt | [82] | |
| Transgenic zebrafish | Hazardous metal pollution in fresh water (Lab scale experiment) | National Taiwan University, Taipei, Taiwan | [83] | |
| Invertebrate | Lamellidens marginalis | Heavy metals: Cr, Mn, Co, Ni, Cu, Zn, Se, As, Sr, Cd, Sn, Sb, Hg, and Pb | Dhimbe reservoir, Maharashtra, India | [84] |
| Adult Odonata (dragonfly) | Monitoring urbanization impact on aquatic environment | Urban streams of Manaus, Amazonas, Brazil | [85] | |
| Mytilus spp. | Microplastic pollution monitoring | Along Norwegian coast, Europe | [86] | |
| Porifera Hymeniacidon perlevis sponge | Cu, Zn, and the hydrocarbon fluoranthene | Along Normandy coast, France | [87] | |
| Nematodes community index | River water polluted by industrial, agricultural and sewage effluents | Beigang River, Taiwan | [88] | |
| Plankton | Marine diatom: (Thalassiosira weissflogii) Estuarine copepod: (Acartia tonsa, Acartia tonsa nauplii) | Cu in pesticide- monitoring in aquatic system | Mondego valley, Figueira da Foz, Portugal | [89] |
| Zooplanktons Chironomus, Oligochaete | Monitoring anthropogenic nitrogen impact on aquatic ecosystem | Lake Nanhu in Wuhan, Hubei Province, China | [90] | |
| Daphnia longispina | S-metolachlor of pesticide- monitoring in aquatic system (lab scale experiment) | Aveiro, Portugal | [91] | |
| Virus | Pepper mild mottle virus (PMMoV), human picobirna-viruses (hPBV), Torque teno virus (TTV) | Human fecal pollution in river | Along Ruhr and Rhine rivers in the North Rhine Westphalia region, Germany | [92] |
| Bacteria | Fecal indicator bacteria (Enterococcus) | Sewage-contaminated groundwater | Avalon Beach, California, USA | [93] |
| Bacteroidales-based biomarkers | Human and livestock wastes | Karst regions in Illinois, Wisconsin, Kentucky, Missouri, USA | [94] | |
| Chromatiaceae, Alcaligenaceae | Palm oil mill effluent (POME) final discharge | River water (approximately 3 km from palm oil mill), Malaysia | [95] | |
| Clostridia, Epsilonproteobacteria | Paper mill effluent | Daling River, Northeast China | [73] | |
| Cupriavidusgilardii, Ralstoniapickettii | Heavy metals (Cu, Zn, Fe and Ni) | Cataño, Puerto Rico | [96] | |
| Bifidobacterium spp. | Point source pollution monitoring such as from agricultural, recreation and water supply | Nanshih River, Taiwan | [44] |
| Illumina Platform/Sequencer | Applications | References |
|---|---|---|
| Illumina HiSeq 2000 | Metagenomic analyses of microbial community pattern in an anaerobic digestion sludge of a full-scale municipal wastewater treatment plant (WWTP). | [147] |
| Illumina HiSeq | Profiling of bacterial community changes in downstream as compared to upstream sections of a river receiving untreated domestic wastewater discharge. | [148] |
| Illumina HiSeq 2500 | Characterization of bacterial community patterns due to seasonal changes in anthropogenically disturbed rivers. | [149] |
| Illumina HiSeq 2500 | Analysis of structures and functions of river water and sediment microbial community during aerobic and anaerobic conditions of endocrine disruptor (17β-estradiol) biodegradation. | [150] |
| Illumina MiSeq | Determination of potential bioindicator from bacterial community obtained from a palm oil mill effluent (POME) treatment system and the receiving waterway. | [95] |
| Illumina MiSeq | Determination of pathogenic bacteria within the bacterial community utilizable for risk assessment of urban surface water. | [151] |
| Illumina MiSeq | A study on the impact of surface water introduction on the microbial community diversity and function within a spring pool in a cave influenced by seasonal changes factor. | [152] |
| Illumina MiSeq | Assessment of the unique presence of targeted bacterial indicator (Alcaligenaceae and Chromatiaceae) within the microbial community in POME final discharge polluted rivers as compared to other non-POME polluted streams. | [153] |
| Sample | Site | Platform | Reference |
|---|---|---|---|
| Activated sludge | Wastewater Treatment Plant (WWTP), Hong Kong | Illumina Hi-Seq2000 | [185] |
| Wastewater wet oxidation effluents | Rovereto, Italy | Illumina Hi-Seq2000 | [179] |
| Deep seafloor sediment | Gulf of Mexico | Illumina Hi-Seq2500 | [186] |
| Deep seawater | Northeast Pacific Ocean | Roche GS FLX Titanium chemistry pyrosequencing | [187] |
| River water | Amazon River, South America | Illumina Hi-Seq2500 | [188] |
| River and seawater | Columbia River, estuary and plume | Illumina Hi-Seq1000 | [189] |
| Biofilm | Tamagawa River, Japan | Illumina MiSeq | [190] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zolkefli, N.; Sharuddin, S.S.; Yusoff, M.Z.M.; Hassan, M.A.; Maeda, T.; Ramli, N. A Review of Current and Emerging Approaches for Water Pollution Monitoring. Water 2020, 12, 3417. https://0-doi-org.brum.beds.ac.uk/10.3390/w12123417
Zolkefli N, Sharuddin SS, Yusoff MZM, Hassan MA, Maeda T, Ramli N. A Review of Current and Emerging Approaches for Water Pollution Monitoring. Water. 2020; 12(12):3417. https://0-doi-org.brum.beds.ac.uk/10.3390/w12123417
Chicago/Turabian StyleZolkefli, Nurhasliza, Siti Suhailah Sharuddin, Mohd Zulkhairi Mohd Yusoff, Mohd Ali Hassan, Toshinari Maeda, and Norhayati Ramli. 2020. "A Review of Current and Emerging Approaches for Water Pollution Monitoring" Water 12, no. 12: 3417. https://0-doi-org.brum.beds.ac.uk/10.3390/w12123417