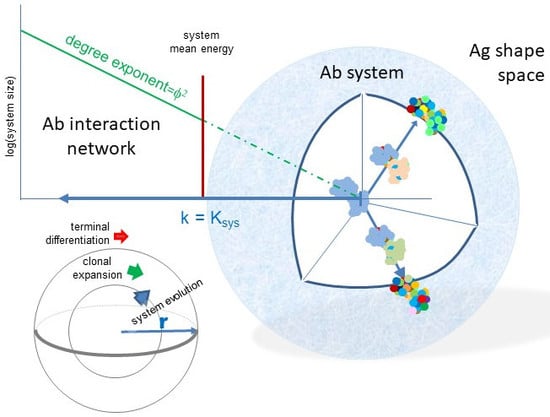
Network Organization of Antibody Interactions in Sequence and Structure Space: the RADARS Model
Abstract
:1. Introduction
2. Antibody Clonal Representation in Sequence Space
3. Antibody Interaction Representation in Structure Space
3.1. Structural Resolution of Molecular Recognition as a Measure of Interaction Strength
3.2. B-Cell Development in Interaction Space
3.3. Immune Responses in Interaction Space
4. Characterization of the Antibody Interaction Network
4.1. Distribution of Binding Energies in the System
4.2. Hierarchy of Binding Events and Geometry of the System
4.3. A Scale-Free Network of Interactions
5. Steps towards Validation and Application of the Model
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Müller, V.; de Boer, R.J.; Bonhoeffer, S.; Szathmáry, E. An evolutionary perspective on the systems of adaptive immunity. Biol Rev. Camb Philos Soc. 2017, 93, 505–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eberl, G. A new vision of immunity: Homeostasis of the superorganism. Mucosal Immunol. 2010, 3, 450–460. [Google Scholar] [CrossRef] [PubMed]
- Sleator, R.D. The human superorganism of microbes and men. Med. Hypotheses 2010, 74, 214–215. [Google Scholar] [CrossRef] [PubMed]
- Pradeu, T.; Carosella, E.D. On the definition of a criterion of immunogenicity. Proc. Natl. Acad. Sci. USA 2006, 103, 17858–17861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, M.M.; Tato, C.M.; Furman, D. Systems immunology: Just getting started. Nat. Immunol. 2017, 18, 725–732. [Google Scholar] [CrossRef]
- Prechl, J. A generalized quantitative antibody homeostasis model: Maintenance of global antibody equilibrium by effector functions. Clin. Transl. Immunol. 2017, 6, e161. [Google Scholar] [CrossRef] [Green Version]
- Boyd, S.D.; Joshi, S.A. High-Throughput DNA Sequencing Analysis of Antibody Repertoires. Microbiol. Spectr. 2014, 2. [Google Scholar] [CrossRef] [Green Version]
- Vergani, S.; Korsunsky, I.; Mazzarello, A.N.; Ferrer, G.; Chiorazzi, N.; Bagnara, D. Novel Method for High-Throughput Full-Length IGHV-D-J Sequencing of the Immune Repertoire from Bulk B-Cells with Single-Cell Resolution. Front. Immunol. 2017, 8, 1157. [Google Scholar] [CrossRef] [Green Version]
- Friedensohn, S.; Lindner, J.M.; Cornacchione, V.; Iazeolla, M.; Miho, E.; Zingg, A.; Meng, S.; Traggiai, E.; Reddy, S.T. Synthetic standards combined with error and bias correction improve the accuracy and quantitative resolution of antibody repertoire sequencing in human naïve and memory B cells. Front. Immunol. 2018, 9, 1401. [Google Scholar] [CrossRef] [Green Version]
- Miho, E.; Yermanos, A.; Weber, C.R.; Berger, C.T.; Reddy, S.T.; Greiff, V. Computational Strategies for Dissecting the High-Dimensional Complexity of Adaptive Immune Repertoires. Front. Immunol. 2018, 9, 224. [Google Scholar] [CrossRef]
- Greiff, V.; Miho, E.; Menzel, U.; Reddy, S.T. Bioinformatic and statistical analysis of adaptive immune repertoires. Trends Immunol. 2015, 36, 738–749. [Google Scholar] [CrossRef]
- Prechl, J.; Papp, K.; Erdei, A. Antigen microarrays: Descriptive chemistry or functional immunomics? Trends Immunol. 2010, 31, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Merbl, Y.; Zucker-Toledano, M.; Quintana, F.J.; Cohen, I.R. Newborn humans manifest autoantibodies to defined self molecules detected by antigen microarray informatics. J. Clin. Investig. 2007, 117, 712–718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bacarese-Hamilton, T.; Bistoni, F.; Crisanti, A. Protein microarrays: From serodiagnosis to whole proteome scale analysis of the immune response against pathogenic microorganisms. BioTechniques 2002, 33, 24–29. [Google Scholar] [CrossRef]
- Wang, J.; Lin, J.; Bardina, L.; Goldis, M.; Nowak-Wegrzyn, A.; Shreffler, W.G.; Sampson, H.A. Correlation of IgE/IgG4 milk epitopes and affinity of milk-specific IgE antibodies with different phenotypes of clinical milk allergy. J. Allergy Clin. Immunol. 2010, 125, 695–702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grötzinger, C. Peptide microarrays for medical applications in autoimmunity, infection, and cancer. Methods Mol. Biol. 2016, 1352, 213–221. [Google Scholar] [CrossRef]
- Sjöberg, R.; Mattsson, C.; Andersson, E.; Hellström, C.; Uhlen, M.; Schwenk, J.M.; Ayoglu, B.; Nilsson, P. Exploration of high-density protein microarrays for antibody validation and autoimmunity profiling. N. Biotechnol. 2016, 33, 582–592. [Google Scholar] [CrossRef]
- Bradford, T.J.; Wang, X.; Chinnaiyan, A.M. Cancer immunomics: Using autoantibody signatures in the early detection of prostate cancer. Urol. Oncol. 2006, 24, 237–242. [Google Scholar] [CrossRef]
- Zantow, J.; Moreira, G.M.S.G.; Dübel, S.; Hust, M. ORFeome Phage Display. Methods Mol. Biol. 2018, 1701, 477–495. [Google Scholar] [CrossRef]
- Wu, F.-L.; Lai, D.-Y.; Ding, H.-H.; Tang, Y.-J.; Xu, Z.-W.; Ma, M.-L.; Guo, S.-J.; Wang, J.-F.; Shen, N.; Zhao, X.-D.; et al. Identification of serum biomarkers for systemic lupus erythematosus using a library of phage displayed random peptides and deep sequencing. Mol. Cell Proteom. 2019, 18, 1851–1863. [Google Scholar] [CrossRef]
- Caetano-Anollés, G.; Wang, M.; Caetano-Anollés, D.; Mittenthal, J.E. The origin, evolution and structure of the protein world. Biochem J. 2009, 417, 621–637. [Google Scholar] [CrossRef] [PubMed]
- Scaviner, D.; Barbié, V.; Ruiz, M.; Lefranc, M.P. Protein displays of the human immunoglobulin heavy, kappa and lambda variable and joining regions. Exp. Clin. Immunogenet. 1999, 16, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, K.; Kelm, S.; Kovaltsuk, A.; Galson, J.D.; Kelly, D.; Trück, J.; Regep, C.; Leem, J.; Wong, W.K.; Nowak, J.; et al. Structurally mapping antibody repertoires. Front. Immunol. 2018, 9, 1698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madi, A.; Kenett, D.Y.; Bransburg-Zabary, S.; Merbl, Y.; Quintana, F.J.; Tauber, A.I.; Cohen, I.R.; Ben-Jacob, E. Network theory analysis of antibody-antigen reactivity data: The immune trees at birth and adulthood. PLoS ONE 2011, 6, e17445. [Google Scholar] [CrossRef] [PubMed]
- Jerne, N.K. Towards a network theory of the immune system. Ann. Immunol. 1974, 125C, 373–389. [Google Scholar]
- Coutinho, A. The network theory: 21 years later. Scand. J. Immunol. 1995, 42, 3–8. [Google Scholar] [CrossRef]
- Miho, E.; Greiff, V.; Roskar, R.; Reddy, S.T. The fundamental principles of antibody repertoire architecture revealed by large-scale network analysis. BioRxiv 2017. [Google Scholar] [CrossRef] [Green Version]
- Prechl, J. A generalized quantitative antibody homeostasis model: Regulation of B-cell development by BCR saturation and novel insights into bone marrow function. Clin. Transl. Immunol. 2017, 6, e130. [Google Scholar] [CrossRef]
- Prechl, J. A generalized quantitative antibody homeostasis model: Antigen saturation, natural antibodies and a quantitative antibody network. Clin. Transl Immunol. 2017, 6, e131. [Google Scholar] [CrossRef]
- Elhanati, Y.; Sethna, Z.; Marcou, Q.; Callan, C.G.; Mora, T.; Walczak, A.M. Inferring processes underlying B-cell repertoire diversity. Philos Trans. R. Soc. Lond. B Biol. Sci. 2015, 370. [Google Scholar] [CrossRef] [Green Version]
- Kovaltsuk, A.; Leem, J.; Kelm, S.; Snowden, J.; Deane, C.M.; Krawczyk, K. Observed Antibody Space: A Resource for Data Mining Next-Generation Sequencing of Antibody Repertoires. J. Immunol. 2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rettig, T.A.; Ward, C.; Bye, B.A.; Pecaut, M.J.; Chapes, S.K. Characterization of the naive murine antibody repertoire using unamplified high-throughput sequencing. PLoS ONE 2018, 13, e0190982. [Google Scholar] [CrossRef] [Green Version]
- Bashford-Rogers, R.J.M.; Palser, A.L.; Huntly, B.J.; Rance, R.; Vassiliou, G.S.; Follows, G.A.; Kellam, P. Network properties derived from deep sequencing of human B-cell receptor repertoires delineate B-cell populations. Genome Res. 2013, 23, 1874–1884. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, S.; Ferrara, F.; Naranjo, L.; Erasmus, M.F.; Hraber, P.; Bradbury, A.R.M. Many Routes to an Antibody Heavy-Chain CDR3: Necessary, Yet Insufficient, for Specific Binding. Front. Immunol. 2018, 9, 395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greiff, V.; Menzel, U.; Miho, E.; Weber, C.; Riedel, R.; Cook, S.; Valai, A.; Lopes, T.; Radbruch, A.; Winkler, T.H.; et al. Systems Analysis Reveals High Genetic and Antigen-Driven Predetermination of Antibody Repertoires throughout B Cell Development. Cell Rep. 2017, 19, 1467–1478. [Google Scholar] [CrossRef] [Green Version]
- DeFalco, J.; Harbell, M.; Manning-Bog, A.; Baia, G.; Scholz, A.; Millare, B.; Sumi, M.; Zhang, D.; Chu, F.; Dowd, C.; et al. Non-progressing cancer patients have persistent B cell responses expressing shared antibody paratopes that target public tumor antigens. Clin. Immunol. 2018, 187, 37–45. [Google Scholar] [CrossRef]
- Bukhari, S.A.C.; O’Connor, M.J.; Martínez-Romero, M.; Egyedi, A.L.; Willrett, D.; Graybeal, J.; Musen, M.A.; Rubelt, F.; Cheung, K.-H.; Kleinstein, S.H. The CAIRR Pipeline for Submitting Standards-Compliant B and T Cell Receptor Repertoire Sequencing Studies to the National Center for Biotechnology Information Repositories. Front. Immunol. 2018, 9, 1877. [Google Scholar] [CrossRef]
- Breden, F.; Luning Prak, E.T.; Peters, B.; Rubelt, F.; Schramm, C.A.; Busse, C.E.; Vander Heiden, J.A.; Christley, S.; Bukhari, S.A.C.; Thorogood, A.; et al. Reproducibility and reuse of adaptive immune receptor repertoire data. Front. Immunol. 2017, 8, 1418. [Google Scholar] [CrossRef]
- Rubelt, F.; Busse, C.E.; Bukhari, S.A.C.; Bürckert, J.-P.; Mariotti-Ferrandiz, E.; Cowell, L.G.; Watson, C.T.; Marthandan, N.; Faison, W.J.; Hershberg, U.; et al. Adaptive Immune Receptor Repertoire Community recommendations for sharing immune-repertoire sequencing data. Nat. Immunol. 2017, 18, 1274–1278. [Google Scholar] [CrossRef]
- Rosenfeld, A.M.; Meng, W.; Luning Prak, E.T.; Hershberg, U. Immunedb, a novel tool for the analysis, storage, and dissemination of immune repertoire sequencing data. Front. Immunol. 2018, 9, 2107. [Google Scholar] [CrossRef] [Green Version]
- Corrie, B.D.; Marthandan, N.; Zimonja, B.; Jaglale, J.; Zhou, Y.; Barr, E.; Knoetze, N.; Breden, F.M.W.; Christley, S.; Scott, J.K.; et al. iReceptor: A platform for querying and analyzing antibody/B-cell and T-cell receptor repertoire data across federated repositories. Immunol Rev. 2018, 284, 24–41. [Google Scholar] [CrossRef] [PubMed]
- Avram, O.; Vaisman-Mentesh, A.; Yehezkel, D.; Ashkenazy, H.; Pupko, T.; Wine, Y. ASAP—A Webserver for Immunoglobulin-Sequencing Analysis Pipeline. Front. Immunol. 2018, 9, 1686. [Google Scholar] [CrossRef] [PubMed]
- Kringelum, J.V.; Nielsen, M.; Padkjær, S.B.; Lund, O. Structural analysis of B-cell epitopes in antibody: Protein complexes. Mol. Immunol. 2013, 53, 24–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wedemayer, G.J.; Patten, P.A.; Wang, L.H.; Schultz, P.G.; Stevens, R.C. Structural insights into the evolution of an antibody combining site. Science 1997, 276, 1665–1669. [Google Scholar] [CrossRef]
- Chen, J.; Sawyer, N.; Regan, L. Protein-protein interactions: General trends in the relationship between binding affinity and interfacial buried surface area. Protein Sci. 2013, 22, 510–515. [Google Scholar] [CrossRef] [Green Version]
- Brooijmans, N.; Sharp, K.A.; Kuntz, I.D. Stability of macromolecular complexes. Proteins 2002, 48, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Derrida, B. Random-energy model: An exactly solvable model of disordered systems. Phys. Rev. B 1981, 24, 2613–2626. [Google Scholar] [CrossRef]
- Mézard, M.; Montanari, A. “The random energy model.” In Information, Physics, and Computation; Oxford University Press: Oxford, UK, 2009; pp. 93–105. [Google Scholar] [CrossRef]
- Onuchic, J.N.; Luthey-Schulten, Z.; Wolynes, P.G. Theory of protein folding: The energy landscape perspective. Annu. Rev. Phys. Chem. 1997, 48, 545–600. [Google Scholar] [CrossRef] [Green Version]
- Childs, L.M.; Baskerville, E.B.; Cobey, S. Trade-offs in antibody repertoires to complex antigens. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370. [Google Scholar] [CrossRef]
- Zheng, X.; Wang, J. The universal statistical distributions of the affinity, equilibrium constants, kinetics and specificity in biomolecular recognition. PLoS Comput. Biol. 2015, 11, e1004212. [Google Scholar] [CrossRef]
- Melchers, F. Checkpoints that control B cell development. J. Clin. Investig. 2015, 125, 2203–2210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, I.R. Biomarkers, self-antigens and the immunological homunculus. J. Autoimmun. 2007, 29, 246–249. [Google Scholar] [CrossRef] [PubMed]
- Tangye, S.G.; Tarlinton, D.M. Memory B cells: Effectors of long-lived immune responses. Eur. J. Immunol. 2009, 39, 2065–2075. [Google Scholar] [CrossRef] [PubMed]
- Shah, H.B.; Smith, K.; Wren, J.D.; Webb, C.F.; Ballard, J.D.; Bourn, R.L.; James, J.A.; Lang, M.L. Insights From Analysis of Human Antigen-Specific Memory B Cell Repertoires. Front. Immunol. 2018, 9, 3064. [Google Scholar] [CrossRef]
- Neuberger, M.S.; Ehrenstein, M.R.; Rada, C.; Sale, J.; Batista, F.D.; Williams, G.; Milstein, C. Memory in the B-cell compartment: Antibody affinity maturation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000, 355, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Good-Jacobson, K.L.; Shlomchik, M.J. Plasticity and heterogeneity in the generation of memory B cells and long-lived plasma cells: The influence of germinal center interactions and dynamics. J. Immunol. 2010, 185, 3117–3125. [Google Scholar] [CrossRef]
- Hammarlund, E.; Thomas, A.; Amanna, I.J.; Holden, L.A.; Slayden, O.D.; Park, B.; Gao, L.; Slifka, M.K. Plasma cell survival in the absence of B cell memory. Nat. Commun. 2017, 8, 1781. [Google Scholar] [CrossRef] [Green Version]
- Batista, F.D.; Neuberger, M.S. Affinity dependence of the B cell response to antigen: A threshold, a ceiling, and the importance of off-rate. Immunity 1998, 8, 751–759. [Google Scholar] [CrossRef] [Green Version]
- Reed, W.J.; Jorgensen, M. The Double Pareto-Lognormal Distribution—A New Parametric Model for Size Distributions. Commun. Stat.—Theory Methods 2004, 33, 1733–1753. [Google Scholar] [CrossRef]
- Mitzenmacher, M. Dynamic models for file sizes and double pareto distributions. Internet Math. 2004, 1, 305–333. [Google Scholar] [CrossRef] [Green Version]
- Monroe, J.G. Ligand-independent tonic signaling in B-cell receptor function. Curr. Opin. Immunol. 2004, 16, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Akkaya, M.; Kwak, K.; Pierce, S.K. B cell memory: Building two walls of protection against pathogens. Nat. Rev. Immunol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.D.; Wilmore, J.R.; Allman, D. Cellular dynamics of memory B cell populations: Igm+ and igg+ memory B cells persist indefinitely as quiescent cells. J. Immunol. 2015, 195, 4753–4759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilmore, J.R.; Allman, D. Here, There, and Anywhere? Arguments for and against the Physical Plasma Cell Survival Niche. J. Immunol. 2017, 199, 839–845. [Google Scholar] [CrossRef]
- Lightman, S.M.; Utley, A.; Lee, K.P. Survival of Long-Lived Plasma Cells (LLPC): Piecing Together the Puzzle. Front. Immunol. 2019, 10, 965. [Google Scholar] [CrossRef]
- Shlomchik, M.J. Do memory B cells form secondary germinal centers? Yes and no. Cold Spring Harb. Perspect Biol. 2018, 10. [Google Scholar] [CrossRef]
- Phan, T.G.; Paus, D.; Chan, T.D.; Turner, M.L.; Nutt, S.L.; Basten, A.; Brink, R. High affinity germinal center B cells are actively selected into the plasma cell compartment. J. Exp. Med. 2006, 203, 2419–2424. [Google Scholar] [CrossRef]
- O’Connor, B.P.; Vogel, L.A.; Zhang, W.; Loo, W.; Shnider, D.; Lind, E.F.; Ratliff, M.; Noelle, R.J.; Erickson, L.D. Imprinting the fate of antigen-reactive B cells through the affinity of the B cell receptor. J. Immunol. 2006, 177, 7723–7732. [Google Scholar] [CrossRef] [Green Version]
- Song, C.; Havlin, S.; Makse, H.A. Origins of fractality in the growth of complex networks. Nat. Phys. 2006, 2, 275–281. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.S.; Goh, K.I.; Kahng, B.; Kim, D. Fractality and self-similarity in scale-free networks. New J. Phys. 2007, 9, 177. [Google Scholar] [CrossRef]
- Albert, R.; Barabási, A.-L. Statistical mechanics of complex networks. Rev. Mod. Phys. 2002, 74, 47–97. [Google Scholar] [CrossRef] [Green Version]
- Barabási, A.-L.; Oltvai, Z.N. Network biology: Understanding the cell’s functional organization. Nat. Rev. Genet. 2004, 5, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Morbach, H.; Eichhorn, E.M.; Liese, J.G.; Girschick, H.J. Reference values for B cell subpopulations from infancy to adulthood. Clin. Exp. Immunol. 2010, 162, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Brieva, J.A.; Roldán, E.; De la Sen, M.L.; Rodriguez, C. Human in vivo-induced spontaneous IgG-secreting cells from tonsil, blood and bone marrow exhibit different phenotype and functional level of maturation. Immunology 1991, 72, 580–583. [Google Scholar]
- Miho, E.; Roškar, R.; Greiff, V.; Reddy, S.T. Large-scale network analysis reveals the sequence space architecture of antibody repertoires. Nat. Commun. 2019, 10, 1321. [Google Scholar] [CrossRef] [Green Version]
- Asti, L.; Uguzzoni, G.; Marcatili, P.; Pagnani, A. Maximum-Entropy Models of Sequenced Immune Repertoires Predict Antigen-Antibody Affinity. PLoS Comput. Biol. 2016, 12, e1004870. [Google Scholar] [CrossRef] [Green Version]
- Marillet, S.; Lefranc, M.-P.; Boudinot, P.; Cazals, F. Novel Structural Parameters of Ig-Ag Complexes Yield a Quantitative Description of Interaction Specificity and Binding Affinity. Front. Immunol. 2017, 8, 34. [Google Scholar] [CrossRef] [Green Version]
- Sirin, S.; Apgar, J.R.; Bennett, E.M.; Keating, A.E. AB-Bind: Antibody binding mutational database for computational affinity predictions. Protein Sci. 2016, 25, 393–409. [Google Scholar] [CrossRef] [Green Version]
- Geng, C.; Xue, L.C.; Roel-Touris, J.; Bonvin, A.M.J.J. Finding the ΔΔG spot: Are predictors of binding affinity changes upon mutations in protein-protein interactions ready for it? WIREs Comput. Mol. Sci. 2019, e1410. [Google Scholar] [CrossRef] [Green Version]
- Burnet, F.M. A modification of Jerne’s theory of antibody production using the concept of clonal selection. CA Cancer J. Clin. 1976, 26, 119–121. [Google Scholar] [CrossRef] [Green Version]
- Farmer, D.J.; Packard, N.; Perelson, A. The immune system, adaptation, and machine learning. Phys. D Nonlinear Phenom. 1986, 22, 187–204. [Google Scholar] [CrossRef]
- Schulz, R.; Werner, B.; Behn, U. Self-tolerance in a minimal model of the idiotypic network. Front. Immunol. 2014, 5, 86. [Google Scholar] [CrossRef] [PubMed]
- Mäkelä, T.; Annila, A. Natural patterns of energy dispersal. Phys. Life Rev. 2010, 7, 477–498. [Google Scholar] [CrossRef] [PubMed]
- Hartonen, T.; Annila, A. Natural networks as thermodynamic systems. Complexity 2012, 18, 53–62. [Google Scholar] [CrossRef]






© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prechl, J. Network Organization of Antibody Interactions in Sequence and Structure Space: the RADARS Model. Antibodies 2020, 9, 13. https://0-doi-org.brum.beds.ac.uk/10.3390/antib9020013
Prechl J. Network Organization of Antibody Interactions in Sequence and Structure Space: the RADARS Model. Antibodies. 2020; 9(2):13. https://0-doi-org.brum.beds.ac.uk/10.3390/antib9020013
Chicago/Turabian StylePrechl, József. 2020. "Network Organization of Antibody Interactions in Sequence and Structure Space: the RADARS Model" Antibodies 9, no. 2: 13. https://0-doi-org.brum.beds.ac.uk/10.3390/antib9020013