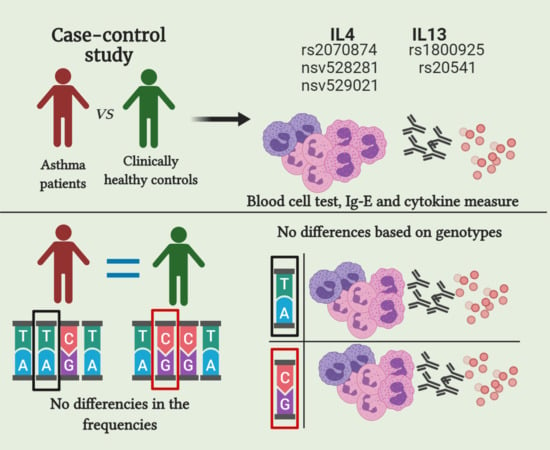
Single Nucleotide and Copy-Number Variants in IL4 and IL13 Are Not Associated with Asthma Susceptibility or Inflammatory Markers: A Case-Control Study in a Mexican-Mestizo Population
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Biologic Samples
2.3. DNA Extraction
2.4. Genotyping of CNVs and SNPs
2.5. Plasma Cytokines Measurements
2.6. Statistical Analysis
3. Results
3.1. Population Description
3.2. Allele and Genotypes Comparison
3.3. Association Analysis Applying Genetics Models
3.4. Linkage Disequilibrium (LD) and Haplotype Block (HB) Construction
3.5. Copy-Number Variation: Identification and Frequency Comparison
3.6. IL-4 and IL-13 Measurement and Analysis
3.7. Correlation between BBS and Clinical Data
3.8. IgE Levels Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Global Strategy for Asthma Management and Prevention Revised 2006. Available online: www.ginasthma.org (accessed on 21 August 2019).
- Anuario 1984–2016. Available online: http://www.epidemiologia.salud.gob.mx/anuario/html/anuarios.html (accessed on 21 August 2019).
- Bousquet, J.; Bousquet, P.J.; Godard, P.; Daures, J.-P. The public health implications of asthma. Bull. World Health Organ. 2005, 83, 548–554. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/16175830 (accessed on 21 August 2019). [PubMed]
- Gilliland, F.D.; Berhane, K.; McConnell, R.; Gauderman, W.J.; Vora, H.; Rappaport, E.B.; Avol, E.; Peters, J.M. Maternal smoking during pregnancy, environmental tobacco smoke exposure and childhood lung function. Thorax 2000, 55, 271–276. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/10722765 (accessed on 21 August 2019). [CrossRef] [PubMed] [Green Version]
- Lødrup Carlsen, K.C.; Jaakkola, J.J.; Nafstad, P.; Carlsen, K.H. In utero exposure to cigarette smoking influences lung function at birth. Eur. Respir. J. 1997, 10, 1774–1779. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/9272918 (accessed on 21 August 2019). [CrossRef] [PubMed] [Green Version]
- Kim, S.H.; Kim, Y.K.; Park, H.W.; Kim, S.H.; Kim, S.H.; Ye, Y.M.; Min, K.U.; Park, H.S. Adenosine deaminase and adenosine receptor polymorphisms in aspirin-intolerant asthma. Respir. Med. 2009, 103, 356–363. Available online: https://0-linkinghub-elsevier-com.brum.beds.ac.uk/retrieve/pii/S0954611108003594 (accessed on 21 August 2019). [CrossRef] [PubMed] [Green Version]
- Kamada, F.; Suzuki, Y.; Shao, C.; Tamari, M.; Hasegawa, K.; Hirota, T.; Shimizu, M.; Takahashi, N.; Mao, X.Q.; Doi, S.; et al. Association of the hCLCA1 gene with childhood and adult asthma. Genes Immun. 2004, 5, 540–547. Available online: http://0-www-nature-com.brum.beds.ac.uk/articles/6364124 (accessed on 21 August 2019). [CrossRef]
- Wang, K.; Feng, Y.L.; Wen, F.Q.; Chen, X.R.; Ou, X.-M.; Xu, D.; Yang, J.; Deng, Z.P. Increased expression of human calcium-activated chloride channel 1 is correlated with mucus overproduction in the airways of Chinese patients with chronic obstructive pulmonary disease. Chin. Med. J. (Engl.) 2007, 120, 1051–1057. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/17637221 (accessed on 21 August 2019). [CrossRef]
- Chan, I.H.S.; Tang, N.L.S.; Leung, T.F.; Lam, Y.Y.O.; Wong, G.W.K.; Wong, C.K.; Lam, C.W.K. Association of plasma soluble CTLA-4 with lung function and GenePolymorphism in Chinese asthmatic children. Int. Arch. Allergy Immunol. 2010, 152, 113–121. [Google Scholar] [CrossRef]
- Tantisira, K.G.; Weiss, S.T. Complex interactions in complex traits: Obesity and asthma. Thorax 2001, 56 (suppl. 2), ii64–ii74. [Google Scholar]
- Moffatt, M.F.; James, A.; Ryan, G.; Musk, A.W.; Cookson, W.O.C.M. Extended tumour necrosis factor/HLA-DR haplotypes and asthma in an Australian population sample. Thorax 1999, 54, 757–761. Available online: http://0-thorax-bmj-com.brum.beds.ac.uk/cgi/doi/10.1136/thx.54.9.757 (accessed on 21 August 2019). [CrossRef] [Green Version]
- Shin, H.D.; Park, B.L.; Kim, L.H.; Jung, J.H.; Wang, H.J.; Kim, Y.J.; Park, H.S.; Hong, S.J.; Choi, B.W.; Kim, D.J.; et al. Association of tumor necrosis factor polymorphisms with asthma and serum total IgE. Hum. Mol. Genet. 2004, 13, 397–403. [Google Scholar] [CrossRef] [Green Version]
- Joubert, P.; Lajoie-Kadoch, S.; Welman, M.; Dragon, S.; Létuvée, S.; Tolloczko, B.; Halayko, A.J.; Gounni, A.S.; Maghni, K.; Hamid, Q. Expression and Regulation of CCR1 by Airway Smooth Muscle Cells in Asthma. J. Immunol. 2008, 180, 1268–1275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, Q.; Moir, L.M.; Black, J.L.; Oliver, B.G.; Burgess, J.K. TGFβ1 induces IL-6 and inhibits IL-8 release in human bronchial epithelial cells: The role of Smad2/3. J. Cell. Physiol. 2010, 225, 846–854. Available online: http://0-doi-wiley-com.brum.beds.ac.uk/10.1002/jcp.22295 (accessed on 21 August 2019). [CrossRef]
- Rose-John, S. Interleukin-6 biology is coordinated by membrane-bound and soluble receptors: Role in inflammation and cancer. J. Leukoc. Biol. 2006, 80, 227–236. [Google Scholar] [CrossRef] [Green Version]
- Yokoyama, A.; Kohno, N.; Fujino, S.; Hamada, H.; Inoue, Y.; Fujioka, S.; Ishida, S.; Hiwada, K. Circulating interleukin-6 levels in patients with bronchial asthma. Am. J. Respir. Crit. Care Med. 1995, 151, 1354–1358. [Google Scholar] [CrossRef] [PubMed]
- Holla, L.I.; Jurajda, M.; Pohunek, P.; Znojil, V. Haplotype analysis of the endothelial nitric oxide synthase gene in asthma. Hum. Immunol. 2008, 69, 306–313. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/18486767 (accessed on 21 August 2019). [CrossRef] [PubMed]
- Hill, M.; Cookson, W.O. A new variant of the beta subunit of the high-affinity receptor for immunoglobulin E (Fc epsilon RI-beta E237G): Associations with measures of atopy and bronchial hyper-responsiveness. Hum. Mol. Genet. 1996, 5, 959–962. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/8817330 (accessed on 21 August 2019). [CrossRef] [PubMed] [Green Version]
- Chai, R.; Liu, B.; Qi, F. The significance of the levels of IL-4, IL-31 and TLSP in patients with asthma and/or rhinitis. Immunotherapy 2017, 9, 331–337. [Google Scholar] [CrossRef]
- Gandhi, N.A.; Pirozzi, G.; Graham, N.M.H. Commonality of the IL-4/IL-13 pathway in atopic diseases. Expert Rev. Clin. Immunol. 2017, 13, 425–437. [Google Scholar] [CrossRef]
- Gour, N.; Wills-Karp, M. IL-4 and IL-13 Signaling in Allergic Airway Disease. Cytokine 2015, 75, 68–78. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.Y.; Kang, B.; Bok, S.H.; Cho, S.S.; Park, D.H. Macmoondongtang modulates Th1-/Th2-related cytokines and alleviates asthma in a murine model. PLoS ONE 2019, 14, e0224517. [Google Scholar] [CrossRef] [Green Version]
- Hunninghake, G.M.; Soto-Quirós, M.E.; Avila, L.; Su, J.; Murphy, A.; Demeo, D.L.; Ly, N.P.; Liang, C.; Sylvia, J.S.; Klanderman, B.J.; et al. Polymorphisms in IL13, total IgE, eosinophilia, and asthma exacerbations in childhood. J. Allergy Clin. Immunol. 2007, 120, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Hansen, S.; Otten, N.D.; Birch, K.; Skovgaard, K.; Hopster-Iversen, C.; Fjeldborg, J. Bronchoalveolar lavage fluid cytokine, cytology and IgE allergen in horses with equine asthma. Vet. Immunol. Immunopathol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Yang, Q.Z.; Wang, W.; Zhang, G.Q.; Yang, J. Increased IL-4- and IL-17-producing CD8+ cells are related to decreased CD39+CD4+Foxp3+ cells in allergic asthma. J. Asthma 2018, 55, 8–14. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/28346024 (accessed on 16 April 2020). [CrossRef]
- Humbert, M.; Corrigan, C.J.; Kimmitt, P.; Till, S.J.; Kay, A.B.; Durham, S.R. Relationship between il-4 and il-5 mrna expression and disease severity in atopic asthma. Am. J. Respir. Crit. Care Med. 1997, 156, 704–708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Gao, X.; Miao, Y.; Wang, Y.; Wang, H.; Cheng, Z.; Wang, X.; Jing, X.; Jia, L.; Dai, L.; et al. NLRP3 regulates macrophage M2 polarization through up-regulation of IL-4 in asthma. Biochem. J. 2018, 475, 1995–2008. [Google Scholar] [CrossRef]
- Ladjemi, M.Z.; Gras, D.; Dupasquier, S.; Detry, B.; Lecocq, M.; Garulli, C.; Fregimilicka, C.; Bouzin, C.; Gohy, S.; Chanez, P.; et al. Bronchial epithelial IgA secretion is impaired in asthma role of IL-4/IL-13. Am. J. Respir. Crit. Care Med. 2018, 197, 1396–1409. [Google Scholar] [CrossRef]
- Xia, F.; Deng, C.; Jiang, Y.; Qu, Y.; Deng, J.; Cai, Z.; Ding, Y.; Guo, Z.; Wang, J. IL4 (interleukin 4) induces autophagy in B cells leading to exacerbated asthma. Autophagy 2018, 14, 450–464. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/29297752 (accessed on 16 April 2020). [CrossRef]
- Agrawal, S.; Townley, R.G. Role of periostin, FENO, IL-13, lebrikzumab, other IL-13 antagonist and dual IL-4/IL-13 antagonist in asthma. Expert Opin. Biol. Ther. 2014, 14, 165–181. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/24283478 (accessed on 16 April 2020). [CrossRef]
- Zhang, F.Q.; Han, X.P.; Zhang, F.; Ma, X.; Xiang, D.; Yang, X.M.; Ou-Yang, H.F.; Li, Z. Therapeutic efficacy of a co-blockade of IL-13 and IL-25 on airway inflammation and remodeling in a mouse model of asthma. Int. Immunopharmacol. 2017, 46, 133–140. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/28282577 (accessed on 16 April 2020). [CrossRef]
- Proboszcz, M.; Paplińska-Goryca, M.; Nejman-Gryz, P.; Górska, K.; Krenke, R. A comparative study of sTREM-1, IL-6 and IL-13 concentration in bronchoalveolar lavage fluid in asthma and COPD: A preliminary study. Adv. Clin. Exp. Med. 2017, 26, 231–236. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/28791839 (accessed on 16 April 2020). [CrossRef] [Green Version]
- Alasandagutti, M.L.; Ansari, M.S.S.; Sagurthi, S.R.; Valluri, V.; Gaddam, S. Role of IL-13 Genetic Variants in Signalling of Asthma. Inflammation 2017, 40, 566–577. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/28083766 (accessed on 16 April 2020). [CrossRef] [PubMed]
- Tang, L.; Lin, H.G.; Chen, B.F. Association of IL-4 promoter polymorphisms with asthma: A meta-analysis. Genet. Mol. Res. 2014, 13, 1383–1394. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/24634237 (accessed on 16 April 2020). [CrossRef] [PubMed]
- Micheal, S.; Minhas, K.; Ishaque, M.; Ahmed, F.; Ahmed, A. IL-4 gene polymorphisms and their association with atopic asthma and allergic rhinitis in Pakistani patients. J. Investig. Allergol. Clin. Immunol. 2013, 23, 107–111. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/23654077 (accessed on 16 April 2020). [PubMed]
- Ricciardolo, F.L.M.; Sorbello, V.; Silvestri, M.; Giacomelli, M.; Debenedetti, V.M.G.; Malerba, M.; Ciprandi, G.; Rossi, G.A.; Rossi, A.; Bontempelli, M. TNF-α, IL-4Rα and IL-4 polymorphisms in mild to severe asthma from Italian Caucasians. Int. J. Immunopathol. Pharmacol. 2013, 26, 75–84. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/23527710 (accessed on 16 April 2020). [CrossRef] [PubMed] [Green Version]
- Li, J.; Lin, L.H.; Wang, J.; Peng, X.; Dai, H.R.; Xiao, H.; Li, F.; Wang, Y.P.; Yang, Z.J.; Li, L. Interleukin-4 and interleukin-13 pathway genetics affect disease susceptibility, serum immunoglobulin e levels, and gene expression in asthma. Ann. Allergy Asthma Immunol. 2014, 113, 173–179. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/24980391 (accessed on 16 April 2020). [CrossRef]
- Wang, R.S.; Jin, H.X.; Shang, S.Q.; Liu, X.Y.; Chen, S.J.; Jin, Z.B. Relación entre la expresión de IL-2 e IL-4 y sus polimorfismos y los riesgos de padecer infección por Mycoplasma pneumoniae y asma en niños. Arch. Bronconeumol. 2015, 51, 571–578. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/25747600 (accessed on 16 April 2020). [CrossRef]
- Xu, Y.; Li, J.; Ding, Z.; Li, J.; Li, B.; Yu, Z.; Tan, W. Association between IL-13 +1923C/T polymorphism and asthma risk: A meta-Analysis based on 26 case-control studies. Biosci. Rep. 2017, 37. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/28057889 (accessed on 16 April 2020). [CrossRef] [Green Version]
- Liu, Z.; Li, P.; Wang, J.; Fan, Q.; Yan, P.; Zhang, X.; Han, B. A meta-analysis of IL-13 polymorphisms and pediatric asthma risk. Med. Sci. Monit. 2014, 20, 2617–2623. [Google Scholar]
- Nie, W.; Liu, Y.; Bian, J.; Li, B.; Xiu, Q. Effects of Polymorphisms -1112C/T and +2044A/G in Interleukin-13 Gene on Asthma Risk: A Meta-Analysis. PLoS ONE 2013, 8, e56065. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/23437086 (accessed on 16 April 2020). [CrossRef]
- Li, T.; Ren, Z.; Deng, Y.; Wang, Y.; Zhou, H. Lack of association between RAD50-IL13 polymorphisms and pediatric asthma susceptibility in Northeastern Han Chinese. J. Asthma 2016, 53, 114–118. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/26365633 (accessed on 16 April 2020). [CrossRef]
- Accordini, S.; Calciano, L.; Bombieri, C.; Malerba, G.; Belpinati, F.; Lo Presti, A.R.; Baldan, A.; Ferrari, M.; Perbellini, L. De Marco, R.An Interleukin 13 polymorphism is associated with symptom severity in adult subjects with ever asthma. PLoS ONE 2016, 11, e0151292. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/26986948 (accessed on 16 April 2020). [CrossRef] [PubMed] [Green Version]
- Mei, Q.; Qu, J. Interleukin-13 +2044 G/A and +1923C/T polymorphisms are associated with asthma susceptibility in Asians. Medecine 2017, 96, e9203. [Google Scholar] [CrossRef] [PubMed]
- Birbian, N.; Singh, J.; Jindal, S.K.; Sobti, R.C. High risk association of IL-4 VNTR polymorphism with asthma in a north indian population. Cytokine 2014, 66, 87–94. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/24491812 (accessed on 16 April 2020). [CrossRef]
- Li, L.; Li, Y.; Zeng, X.C.; Li, J.; Du, X.Y. Role of interleukin-4 genetic polymorphisms and environmental factors in the risk of asthma in children. Genet. Mol. Res. 2016, 15, 534–543. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/27819719 (accessed on 16 April 2020). [CrossRef] [PubMed]
- Morales, E.; Duffy, D. Genetics and Gene-Environment Interactions in Childhood and Adult Onset Asthma. Front. Pediatr. 2019, 7, 499. [Google Scholar] [CrossRef] [Green Version]
- Bouzigon, E.; Nadif, R.; Le Moual, N.; Dizier, M.H.; Aschard, H.; Boudier, A.; Bousquet, J.; Chanoine, S.; Donnay, C.; Dumas, O.; et al. Genetic and environmental factors of asthma and allergy: Results of the EGEA study. Rev. Mal. Respir. 2015, 32, 822–840. [Google Scholar] [CrossRef]
- Bråbäck, L.; Lodge, C.J.; Lowe, A.J.; Dharmage, S.C.; Olsson, D.; Forsberg, B. Childhood asthma and smoking exposures before conception—A three-generational cohort study. Pediatr. Allergy Immunol. 2018, 29, 361–368. [Google Scholar] [CrossRef]
- Little, J.; Higgins, J.P.T.; Ioannidis, J.P.A.; Moher, D.; Gagnon, F.; Von Elm, E.; Khoury, M.J.; Cohen, B.; Davey-Smith, G.; Grimshaw, J.; et al. STrengthening the REporting of Genetic Association studies (STREGA)—an extension of the STROBE statement. Eur. J. Clin. Investig. 2009, 39, 247–266. [Google Scholar] [CrossRef] [Green Version]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. Available online: http://0-linkinghub-elsevier-com.brum.beds.ac.uk/retrieve/pii/S0002929707613524 (accessed on 19 October 2019). [CrossRef] [Green Version]
- Henschke, H. De Finetti Diagram. 2008. Available online: https://web.archive.org/web/20110719102655/https://finetti.meb.uni-bonn.de/download (accessed on 24 March 2020).
- Epi InfoTM|CDC. Available online: https://www.cdc.gov/epiinfo/index.html (accessed on 19 February 2018).
- RStudio Team. RStudio: Integrated Development for R; RStudio: Boston, MA, USA, 2015. [Google Scholar]
- Ramírez-Venegas, A.; Velázquez-Uncal, M.; Pérez-Hernández, R.; Guzmán-Bouilloud, N.E.; Falfán-Valencia, R.; Mayar-Maya, M.E.; Aranda-Chávez, A.; Sansores, R.H. Prevalence of COPD and respiratory symptoms associated with biomass smoke exposure in a suburban area. Int. J. COPD 2018, 13, 1727–1734. Available online: https://www.dovepress.com/prevalence-of-copd-and-respiratory-symptoms-associated-with-biomass-sm-peer-reviewed-article-COPD (accessed on 21 November 2018). [CrossRef] [Green Version]
- Kirenga, B.J.; De Jong, C.; Katagira, W.; Kasozi, S.; Mugenyi, L.; Boezen, M.; van der Molen, T.; Kamya, M.R. Prevalence and factors associated with asthma among adolescents and adults in Uganda: A general population based survey. BMC Public Health 2019, 19, 227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oluwole, O.; Arinola, G.O.; Huo, D.; Olopade, C.O. Biomass fuel exposure and asthma symptoms among rural school children in Nigeria. J. Asthma. 2017, 54, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Babalik, A.; Bakirci, N.; Taylan, M.; Bostan, L.; Kiziltaş, Ş.; Başbuğ, Y.; Çalişir, H.C. Biomass smoke exposure as a serious health hazard for women. Tuberk. Toraks 2013, 61, 115–121. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/23875589 (accessed on 31 October 2018). [CrossRef] [PubMed] [Green Version]
- Kodgule, R.; Salvi, S. Exposure to biomass smoke as a cause for airway disease in women and children. Curr. Opin. Allergy Clin. Immunol. 2012, 12, 82–90. [Google Scholar] [CrossRef]
- Noonan, C.W.; Ward, T.J. Asthma randomized trial of indoor wood smoke (ARTIS): Rationale and methods. Contemp. Clin. Trials 2012, 33, 1080–1087. [Google Scholar] [CrossRef] [Green Version]
- Scherzer, R.; Grayson, M.H. Heterogeneity and the origins of asthma. Ann. Allergy Asthma Immunol. 2018, 121, 400–405. [Google Scholar] [CrossRef]
- Gerald, L.B.; Gerald, J.K.; Gibson, L.; Patel, K.; Zhang, S.; McClure, L.A. Changes in environmental tobacco smoke exposure and asthma morbidity among urban school children. Chest 2009, 135, 911–916. [Google Scholar] [CrossRef] [Green Version]
- Levy, L.C.N.; Humarán, I.M.; Arcos, M.A.Á.; Andrade, M.A.R.; Lazcano-Ponce, E.; Hernández-Ávila, M. Encuesta Global de Tabaquismo en Adultos. Available online: https://www.who.int/tobacco/surveillance/survey/gats/mex-report-2015-spanish.pdf (accessed on 21 November 2018).
- Jacobsen, E.A.; Zellner, K.R.; Colbert, D.; Lee, N.A.; Lee, J.J. Eosinophils Regulate Dendritic Cells and Th2 Pulmonary Immune Responses following Allergen Provocation. J. Immunol. 2011, 187, 6059–6068. [Google Scholar] [CrossRef] [Green Version]
- Matucci, A.; Maggi, E.; Vultaggio, A. Eosinophils, the IL-5/IL-5Rα axis, and the biologic effects of benralizumab in severe asthma. Respir. Med. 2019, 160, 105819. [Google Scholar] [CrossRef]
- Schuliga, M. NF-kappaB Signaling in Chronic Inflammatory Airway Disease. Biomolecules 2015, 5, 1266–1283. Available online: https://0-www-mdpi-com.brum.beds.ac.uk/2218-273X/5/3/1266/ (accessed on 24 March 2020). [CrossRef]
- Gibson, P.G.; Simpson, J.L.; Saltos, N. Heterogeneity of airway inflammation in persistent asthma: Evidence of neutrophilic inflammation and increased sputum interleukin-8. Chest 2001, 119, 1329–1336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wenzel, S.E.; Schwartz, L.B.; Langmack, E.L.; Halliday, J.L.; Trudeau, J.B.; Gibbs, R.L.; Chu, H.W. Evidence that severe asthma can be divided pathologically into two inflammatory subtypes with distinct physiologic and clinical characteristics. Am. J. Respir. Crit. Care Med. 1999, 160, 1001–1008. [Google Scholar] [CrossRef] [PubMed]
- Sur, S.; Crotty, T.B.; Kephart, G.M.; Hyma, B.A.; Colby, T.V.; Reed, C.E.; Hunt, L.W.; Gleich, G.J. Sudden-onset fatal asthma: A distinct entity with few eosinophils and relatively more neutrophils in the airway submucosa? Am. Rev. Respir. Dis. 1993, 148, 713–719. [Google Scholar] [CrossRef] [PubMed]
- McLeod, J.J.A.; Baker, B.; Ryan, J.J. Mast cell production and response to IL-4 and IL-13. Cytokine 2015, 75, 57–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panettieri, R.A. Neutrophilic and Pauci-immune Phenotypes in Severe Asthma. Immunol. Allergy Clin. 2016, 36, 569–579. [Google Scholar] [CrossRef]
- Ray, A.; Kolls, J.K. Neutrophilic Inflammation in Asthma and Association with Disease Severity. Trends Immunol. 2017, 38, 942–954. [Google Scholar] [CrossRef]
- Kim, Y.-M.; Kim, H.; Lee, S.; Kim, S.; Lee, J.-U.; Choi, Y.; Park, H.W.; You, G.; Kang, H.; Lee, S.; et al. Airway G-CSF identifies neutrophilic inflammation and contributes to asthma progression. Eur. Respir. J. 2019, 55, 1900827. [Google Scholar] [CrossRef]
- Van Der Velden, V.H.J. Glucocorticoids: Mechanisms of action and anti-inflammatory potential in asthma. Mediat. Inflamm. 1998, 7, 229–237. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/9792333 (accessed on 17 April 2020). [CrossRef]
- Simons, F.E. Benefits and risks of inhaled glucocorticoids in children with persistent asthma. J. Allergy Clin. Immunol. 1998, 102, S77–S84. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/9819313 (accessed on 17 April 2020). [CrossRef]
- Jiang, H.; Zhang, X.; Chi, X.; Wang, J.; Wang, J.; Dou, J. The effect of inhaled glucocorticoid therapy on serum proteomics of asthmatic patients. Chin. J. Tuberc. Respir. Dis. 2014, 37, 274–278. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/24969716 (accessed on 17 April 2020).
- Spahn, J.D. Combination inhaled glucocorticoid/long-acting beta-agonist safety: The long and winding road. Ann. Allergy Asthma Immunol. 2018, 121, 428–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horvath, G.; Vasas, S.; Wanner, A. Inhaled corticosteroids reduce asthma-associated airway hyperperfusion through genomic and nongenomic mechanisms. Pulm. Pharmacol. Ther. 2007, 20, 157–162. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/16765074 (accessed on 17 April 2020). [CrossRef] [PubMed] [Green Version]
- Cornudella, R. Inhaled and systemic corticosteroids in the treatment of asthma. Allergol. Immunopathol. 1994, 22, 217. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/7840023 (accessed on 17 April 2020).
- Heuser, W.; Menaik, R.; Gupta, S.; Rucco, R. Corticosteroid Faceoff: Which is best for treating asthma and COPD exacerbations? JEMS 2017, 42, 19–21. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/29227587 (accessed on 17 April 2020). [PubMed]
- Perret, J.L.; Abramson, M.J. Biomass smoke COPD: A phenotype or a different disease? Respirology 2018, 23, 124–125. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/29215182 (accessed on 12 July 2019). [CrossRef] [PubMed] [Green Version]
- Pérez-Bautista, O.; Montaño, M.; Pérez-Padilla, R.; Zúñiga-Ramos, J.; Camacho-Priego, M.; Barrientos-Gutiérrez, T.; Buendía-Roldan, I.; Velasco-Torres, Y.; Ramos, C. Women with COPD by biomass show different serum profile of adipokines, incretins, and peptide hormones than smokers. Respir. Res. 2018, 19, 239. [Google Scholar] [CrossRef]
- Bui, D.S.; Burgess, J.A.; Matheson, M.C.; Erbas, B.; Perret, J.; Morrison, S.; Giles, G.G.; Hopper, J.L.; Thomas, P.S.; Markos, J.; et al. Ambient wood smoke, traffic pollution and adult asthma prevalence and severity. Respirology 2013, 18, 1101–1107. [Google Scholar] [CrossRef]
- Ocakli, B.; Acarturk, E.; Aksoy, E.; Gungor, S.; Ciyiltepe, F.; Oztas, S.; Oztas, S.; Ozmen, I.; Agca, M.C.; Salturk, C.; et al. The impact of exposure to biomass smoke versus cigarette smoke on inflammatory markers and pulmonary function parameters in patients with chronic respiratory failure. Int. J. COPD 2018, 13, 1261–1267. [Google Scholar] [CrossRef] [Green Version]
- Uddin, M.; Watz, H.; Malmgren, A.; Pedersen, F. NETopethic inflammation in chronic obstructive pulmonary disease and severe asthma. Front. Immunol. 2019, 10, 47. [Google Scholar] [CrossRef]
- Lachowicz-Scroggins, M.E.; Dunican, E.M.; Charbit, A.R.; Raymond, W.; Looney, M.R.; Peters, M.C.; Gordon, E.D.; Woodruff, P.G.; Lefrançais, E.; Phillips, B.R.; et al. Extracellular DNA, neutrophil extracellular traps, and inflammasome activation in severe asthma. Am. J. Respir. Crit. Care Med. 2019, 199, 1076–1085. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/30888839 (accessed on 19 April 2020). [CrossRef]
- Pham, D.L.; Ban, G.Y.; Kim, S.H.; Shin, Y.S.; Ye, Y.M.; Chwae, Y.J.; Park, H.S. Neutrophil autophagy and extracellular DNA traps contribute to airway inflammation in severe asthma. Clin. Exp. Allergy 2017, 47, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Moore, W.C.; Hastie, A.T.; Li, X.; Li, H.; Busse, W.W.; Jarjour, N.N.; Wenzel, S.E.; Peters, S.P.; Meyers, D.A.; Bleecker, E.R.; et al. Sputum neutrophil counts are associated with more severe asthma phenotypes using cluster analysis. J. Allergy Clin. Immunol. 2014, 133, 1557–1563. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/24332216 (accessed on 25 April 2020). [CrossRef] [PubMed] [Green Version]
- Matsunaga, M.C.; Yamauchi, P.S. IL-4 and IL-13 inhibition in atopic dermatitis. J. Drugs Dermatol. 2016, 15, 925–929. [Google Scholar] [PubMed]
- Manson, M.L.; Säfholm, J.; James, A.; Johnsson, A.-K.; Bergman, P.; Al-Ameri, M.; Orre, A.C.; Mårdh, C.K.; Dahlén, S.E.; Adner, M. IL-13 and IL-4, but not IL-5 nor IL-17A, induce hyperresponsiveness in isolated human small airways. J. Allergy Clin. Immunol. 2019. Available online: https://0-linkinghub-elsevier-com.brum.beds.ac.uk/retrieve/pii/S0091674919316185 (accessed on 11 December 2019). [CrossRef] [PubMed] [Green Version]





| Gene | SNP/CNV | Protein | Chr | Position (Bp) | Nucleotide Change | Biological Effect | MAF |
|---|---|---|---|---|---|---|---|
| IL13 | rs20541 | IL-13 | 5 | 132,660,272 | [G/A] | Missense variant | 0.27 |
| rs1800925 | 132,657,117 | [C/T] | Non-coding transcript | 0.25 | |||
| IL4 | rs2070874 | IL-4 | 132,674,018 | [T/C] | 5′UTR variant | 0.4 | |
| IL4 | nsv528281 | IL-4 | 5 | 132,650,771 | 31,482 bp | Insertion | ND |
| nsv529021 | 132,682,524 | 67,809 bp | Deletion | ND |
| Variables | AP | CHS | p |
|---|---|---|---|
| n = 141 | n = 345 | ||
| Demographical Data | |||
| Age (years) | 40.0 (29.0–53.0) | 40 (30.0–51.5) | 0.18 |
| Sex (M/F), % | 32.6/67.4 | 57.9/42.3 | <0.01 |
| Weight (Kg) | 68.0 (62.0–78.7) | 76.8 (68.0–87.2) | <0.01 |
| Height (m) | 1.58 (1.52–1.65) | 1.64 (1.57–1.70) | <0.01 |
| BMI | 27.5 (24.9–31.2) | 28.9 (25.7–32.0) | 0.08 |
| Pharmacological treatment (%) | 97 (68.8%) | NA | NA |
| Corticosteroids (%) | 42 (43.3%) | NA | NA |
| Glucocorticoids (%) | 9 (9.3%) | NA | NA |
| Exposure Data | |||
| Exposed to BBS (%) | 67 (47.5) | 72 (20.9) | <0.01 |
| Years of exposure to BBS | 13.5 (7.5–17.0) | 8.0 (5.0–16.5) | <0.01 |
| Smokers (total, %) | 68 (48.2) | 151 (43.7) | 0.42 |
| Current (%) | 18 (26.5%) | 80 (52.9) | <0.01 |
| Former (%) | 50 (73.5%) | 71 (47.1) | <0.01 |
| Cigarettes per day | 3 (1–10) | 2 (1–6) | 0.44 |
| Years of smoking | 5 (2–11) | 5 (2–15) | 0.15 |
| Never smoker | 73 (51.8) | 194 (56.7) | 0.27 |
| Lung Function Data | |||
| FEV1 * (%) | 38.5 (28.5–67.8) | NA | NA |
| FVC * (%) | 54.5 (46.5–134) | NA | NA |
| FEV1/FVC * (%) | 58.2 (49.0–68.8) | NA | NA |
| FEV1 **(%) | 48.0 (42.8–67.0) | NA | NA |
| FVC ** (%) | 74.5 (57.0–85.8) | NA | NA |
| FEV1/FVC ** (%) | 65.5 (56.8–82.5) | NA | NA |
| Reversibility (%) | 19.2 (15.5–25.3) | NA | NA |
| Blood Cell Counting and Routine Biomarkers | |||
| Leucocytes (%) | 10.7 (8.5–13.3) | 6.7 (5.8–7.6) | <0.01 |
| Neutrophils (%) | 84.8 (72.9–91.6) | 57.6 (51.9–62.0) | <0.01 |
| Lymphocytes (%) | 8.4 (5.6–16.7) | 32.1 (27.4–36.4) | <0.01 |
| Eosinophils (%) | 0.8 (0.1–2.8) | 2.4 (1.6–3.4) | <0.01 |
| Basophils (%) | 3.0 (1.0–4.0) | 3.0 (2.0–4.0) | 0.02 |
| Hemoglobin (g/dL) | 15.2 (13.8–16.1) | 16.1 (14.8–16.9) | <0.01 |
| MCV | 90.7 (87.0–93.3) | 89.8 (87.3–92.4) | 0.34 |
| MCH | 30.0 (29.0–31.0) | 30.0 (28.5–31.5) | 0.57 |
| SNP | Allele | OH | EH | p |
|---|---|---|---|---|
| rs20541 | A | 0.46 | 0.46 | 1.00 |
| rs1800925 | T | 0.38 | 0.43 | 0.06 |
| rs2070874 | C | 0.53 | 0.50 | 0.39 |
| Gene/SNP | AP | CHS | p | OR | CI 95% |
|---|---|---|---|---|---|
| n = 141 (%) | n = 345 (%) | ||||
| IL13 rs20541 | |||||
| GG | 59 (41.84) | 145 (42.03) | 1.00 (Ref) | ||
| GA | 67 (47.52) | 157 (45.51) | 0.81 | 1.05 | (0.69–1.59) |
| AA | 15 (10.64) | 43 (12.46) | 0.86 | (0.44–1.66) | |
| G | 185 (65.60) | 447 (64.78) | 0.82 | 1.04 | (0.78–1.39) |
| A | 97 (34.40) | 243 (35.22) | 0.97 | (0.72–1.29) | |
| IL13 rs1800925 | |||||
| CC | 66 (46.81) | 155 (50.72) | 1.00 (Ref) | ||
| CT | 60 (42.55) | 149 (37.39) | 0.64 | 0.95 | (0.62–1.43) |
| TT | 15 (10.64) | 41 (11.88) | 0.86 | (0.45–1.66) | |
| C | 192 (68.09) | 459 (69.42) | 0.94 | 0.98 | (0.73–1.32) |
| T | 90 (31.91) | 211 (30.58) | 1.02 | (0.76–1.37) | |
| IL4 rs2070874 | |||||
| TT | 36 (25.5) | 90 (26.09) | 1.00 (Ref) | ||
| TC | 68 (48.66) | 181 (52.46) | 0.44 | 0.94 | (0.58–1.51) |
| CC | 37 (26.24) | 74 (21.44) | 1.25 | (0.72–2.17) | |
| T | 140 (49.64) | 361 (52.32) | 0.48 | 0.89 | (0.68–1.19 |
| C | 142 (50.35) | 329 (47.68) | 1.11 | (0.84–1.47) | |
| Model | Genotype | AP | CHS | p | OR | CI 95% |
|---|---|---|---|---|---|---|
| n = 141 (%) | n = 345 (%) | |||||
| IL13 rs20541 | ||||||
| Full-Genotype | GG | 59 (41.84) | 145 (42.03) | 1.00 | 0.99 | (0.67–1.47) |
| GA | 67 (47.52) | 157 (45.51) | 0.69 | 1.08 | (0.73–1.61) | |
| AA | 15 (10.64) | 43 (12.46) | 0.65 | 0.84 | (0.45–1.56) | |
| Dominant | GG | 59 (41.84) | 145 (42.03) | 1.00 | 0.99 | (0.67–1.48) |
| GA + AA | 82 (58.16) | 200 (57.97) | 1.01 | (0.68-1.49) | ||
| Recessive | GG + GA | 126 (89.63) | 302 (87.54) | 0.64 | 1.12 | (0.64–2.23) |
| AA | 15 (10.64) | 43 (12.46) | 0.84 | (0.45–1.56) | ||
| IL13 rs1800925 | ||||||
| Full-Genotype | CC | 66 (46.81) | 155 (50.72) | 0.76 | 1.08 | (0.73–1.59) |
| CT | 60 (42.55) | 149 (37.39) | 0.92 | 0.97 | (0.66–1.45) | |
| TT | 15 (10.64) | 41 (11.88) | 0.76 | 0.88 | (0.47–1.65) | |
| Dominant | CC | 66 (46.81) | 155 (50.72) | 0.76 | 1.08 | (0.73–1.59) |
| CT + TT | 75 (53.19) | 190 (49.27) | 0.93 | (0.63–1.73) | ||
| Recessive | CC + CT | 126 (89.36) | 304 (88.11) | 0.75 | 1.13 | (0.61–2.12) |
| TT | 15 (10.64) | 41 (11.88) | 0.88 | (0.47–1.65) | ||
| IL4 rs2070874 | ||||||
| Full-Genotype | TT | 36 (25.50) | 90 (26.09) | 1.00 | 0.97 | (0.62–1.52) |
| TC | 68 (48.66) | 181 (52.46) | 0.42 | 0.84 | (0.57–1.25) | |
| CC | 37 (26.24) | 74 (21.44) | 0.28 | 1.30 | (0.83-2.05) | |
| Dominant | TT | 36 (25.5) | 90 (26.09) | 1.00 | 0.97 | (0.62–1.52) |
| TC + CC | 105 (74.5) | 255 (73.91) | 1.03 | (0.66–1.61) | ||
| Recessive | TT + TC | 104 (73.76) | 271 (78.56) | 0.28 | 0.77 | (0.49–1.21) |
| CC | 37 (26.24) | 74 (21.44) | 1.30 | (0.87–2.05) | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ambrocio-Ortiz, E.; Galicia-Negrete, G.; Pérez-Rubio, G.; Escobar-Morales, A.J.; Abarca-Rojano, E.; Del Angel-Pablo, A.D.; Castillejos-López, M.D.J.; Falfán-Valencia, R. Single Nucleotide and Copy-Number Variants in IL4 and IL13 Are Not Associated with Asthma Susceptibility or Inflammatory Markers: A Case-Control Study in a Mexican-Mestizo Population. Diagnostics 2020, 10, 273. https://0-doi-org.brum.beds.ac.uk/10.3390/diagnostics10050273
Ambrocio-Ortiz E, Galicia-Negrete G, Pérez-Rubio G, Escobar-Morales AJ, Abarca-Rojano E, Del Angel-Pablo AD, Castillejos-López MDJ, Falfán-Valencia R. Single Nucleotide and Copy-Number Variants in IL4 and IL13 Are Not Associated with Asthma Susceptibility or Inflammatory Markers: A Case-Control Study in a Mexican-Mestizo Population. Diagnostics. 2020; 10(5):273. https://0-doi-org.brum.beds.ac.uk/10.3390/diagnostics10050273
Chicago/Turabian StyleAmbrocio-Ortiz, Enrique, Gustavo Galicia-Negrete, Gloria Pérez-Rubio, Areli J. Escobar-Morales, Edgar Abarca-Rojano, Alma D. Del Angel-Pablo, Manuel D. J. Castillejos-López, and Ramcés Falfán-Valencia. 2020. "Single Nucleotide and Copy-Number Variants in IL4 and IL13 Are Not Associated with Asthma Susceptibility or Inflammatory Markers: A Case-Control Study in a Mexican-Mestizo Population" Diagnostics 10, no. 5: 273. https://0-doi-org.brum.beds.ac.uk/10.3390/diagnostics10050273