The First Yarrowia lipolytica Yeast Models Expressing Hepatitis B Virus X Protein: Changes in Mitochondrial Morphology and Functions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemical Reagents
2.2. Cell Cultures
2.3. Plasmid and Yeast Strain Construction
2.4. Western Blotting
2.5. Mitochondria Visualization in Cells by SIM Microscopy
2.6. Oxidative Stress and Cell Death of Y. lipolytica Cells
2.7. Isolation of Y. lipolytica Mitochondria
2.8. Monitoring of Oxygen Consumption by Yeast Cells and Mitochondria
2.9. Assessment of the Mitochondrial Membrane Potential
2.10. Monitoring of Mitochondrial Swelling
2.11. Assay of ATP Synthesis by Mitochondria
2.12. Assessment of Hydrogen Peroxide Production by Mitochondria
2.13. Mitochondrial Protein Assay
2.14. Statistical Analysis
3. Results
3.1. Creation and Primary Characterization of Yeast HBx-Expressing Cells
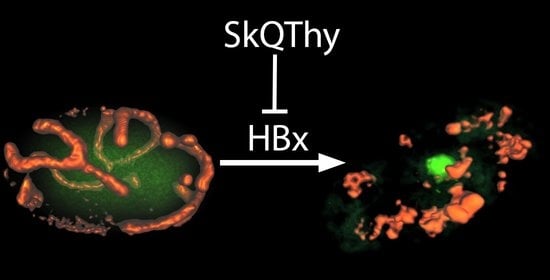
3.2. Morphology of Mitochondria in Yeast Cells
3.3. Oxidative Stress and Cell Death in Yeast Cells
3.4. Energy Parameters of Mitochondria Isolated from Yeast Cells
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, C.; Liu, W.; Zhao, X.; Zhao, L.; Wang, F. Downregulation of HBx Restrains Proliferation, Migration, and Invasion of HepG2 Cells. Anal. Cell. Pathol. (Amst.) 2021, 2021, 6615979. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fu, Y.; Hu, X.; Sun, L.; Tang, D.; Li, N.; Peng, F.; Fan, X.G. The HBx-CTTN interaction promotes cell proliferation and migration of hepatocellular carcinoma via CREB1. Cell Death Dis. 2019, 10, 405. [Google Scholar] [CrossRef] [PubMed]
- Sivasudhan, E.; Blake, N.; Lu, Z.; Meng, J.; Rong, R. Hepatitis B Viral Protein HBx and the Molecular Mechanisms Modulating the Hallmarks of Hepatocellular Carcinoma: A Comprehensive Review. Cells 2022, 11, 741. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Gu, S.; Liang, J.; Lin, Z.; Zheng, S.; Yan, J. The Biological Function of Hepatitis B Virus X Protein in Hepatocellular Carcinoma. Oncol. Res. 2019, 27, 509–514. [Google Scholar] [CrossRef]
- Chen, Z.; Yu, W.; Zhou, Q.; Zhang, J.; Jiang, H.; Hao, D.; Wang, J.; Zhou, Z.; He, C.; Xiao, Z. A Novel lncRNA IHS Promotes Tumor Proliferation and Metastasis in HCC by Regulating the ERK- and AKT/GSK-3β-Signaling Pathways. Mol. Ther. Nucleic Acids 2019, 16, 707–720. [Google Scholar] [CrossRef]
- Giraud, J.; Chalopin, D.; Blanc, J.F.; Saleh, M. Hepatocellular Carcinoma Immune Landscape and the Potential of Immunotherapies. Front. Immunol. 2021, 12, 655697. [Google Scholar] [CrossRef]
- Hu, Z.; Huang, P.; Yan, Y.; Zhou, Z.; Wang, J.; Wu, G. Hepatitis B virus X protein related lncRNA WEE2-AS1 promotes hepatocellular carcinoma proliferation and invasion. Biochem. Biophys. Res. Commun. 2019, 508, 79–86. [Google Scholar] [CrossRef]
- Villanueva, A. Hepatocellular Carcinoma. N. Engl. J. Med. 2019, 380, 1450–1462. [Google Scholar] [CrossRef]
- Zheng, Y.; Ming, P.; Zhu, C.; Si, Y.; Xu, S.; Chen, A.; Wang, J.; Zhang, B. Hepatitis B virus X protein-induced SH2 domain-containing 5 (SH2D5) expression promotes hepatoma cell growth via an SH2D5-transketolase interaction. J. Biol. Chem. 2019, 294, 4815–4827. [Google Scholar] [CrossRef]
- Chou, R.H.; Lee, C.Y.; Chong, L.W.; Chang, K.H.; Lin, C.L.; Yan, K.S.; Tsou, C.; Hsu, Y.C. HBV infection increases the risk of macular degeneration: The roles of HBx-mediated sensitization of retinal pigment epithelial cells to UV and blue light irradiation. J. Transl. Med. 2018, 16, 221. [Google Scholar] [CrossRef] [Green Version]
- Oura, K.; Morishita, A.; Tani, J.; Masaki, T. Tumor Immune Microenvironment and Immunosuppressive Therapy in Hepatocellular Carcinoma: A Review. Int. J. Mol. Sci. 2021, 22, 5801. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.C.; Wu, D.W.; Lin, Y.Y.; Lin, P.L.; Lee, H. Hepatitis B virus X protein represses LKB1 expression to promote tumor progression and poor postoperative outcome in hepatocellular carcinoma. Surgery 2018, 163, 1040–1046. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Liang, H.; Wang, H.; Duan, C.; Yazdani, H.; Zhou, J.; Pan, Y.; Shan, B.; Su, Z.; Wei, J.; et al. Hepatitis B virus-X protein regulates high mobility group box 1 to promote the formation of hepatocellular carcinoma. Oncol. Lett. 2018, 16, 4418–4426. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.X.; Ling, Y.; Wang, H.Y. Role of nonresolving inflammation in hepatocellular carcinoma development and progression. NPJ Precis. Oncol. 2018, 2, 6. [Google Scholar] [CrossRef] [PubMed]
- Njei, B.; Rotman, Y.; Ditah, I.; Lim, J.K. Emerging trends in hepatocellular carcinoma incidence and mortality. Hepatology 2015, 61, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Bautista, W.W.; Osiowy, C.; Klein, J.; Minuk, G.Y. Hepatitis B Virus Infection of Normal Hepatic Stem/Progenitor Cells. J. Clin. Exp. Hepatol. 2019, 9, 34–42. [Google Scholar] [CrossRef]
- Tian, J.H.; Liu, W.D.; Zhang, Z.Y.; Tang, L.H.; Li, D.; Tian, Z.J.; Lin, S.W.; Li, Y.J. Influence of miR-520e-mediated MAPK signalling pathway on HBV replication and regulation of hepatocellular carcinoma cells via targeting EphA2. J. Viral Hepat. 2019, 26, 496–505. [Google Scholar] [CrossRef]
- Ruan, P.; Dai, X.; Sun, J.; He, C.; Huang, C.; Zhou, R.; Cao, Z.; Ye, L. Different types of viral-host junction found in HBV integration breakpoints in HBV-infected patients. Mol. Med. Rep. 2019, 19, 1410–1416. [Google Scholar] [CrossRef]
- An, P.; Xu, J.; Yu, Y.; Winkler, C.A. Host and Viral Genetic Variation in HBV-Related Hepatocellular Carcinoma. Front. Genet. 2018, 9, 261. [Google Scholar] [CrossRef]
- Tu, W.; Gong, J.; Tian, D.; Wang, Z. Hepatitis B Virus X Protein Induces SATB1 Expression Through Activation of ERK and p38MAPK Pathways to Suppress Anoikis. Dig. Dis. Sci. 2019, 64, 3203–3214. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Luo, S.; Xiong, Y. The Variability of Amino Acid Sequences in Hepatitis B Virus. Virol. Sin. 2019, 34, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Zhao, L.; Yuan, Y.; Yun, H.; Zheng, W.; Geng, Y.; Yang, G.; Wang, Y.; Zhao, M.; Zhang, X. HBx represses WDR77 to enhance HBV replication by DDB1-mediated WDR77 degradation in the liver. Theranostics 2021, 11, 8362–8378. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, V.K.; Singh, A.; Dubey, S.K.; Hetta, H.F.; John, J.; Singh, M.P. Molecular mechanistic insight of hepatitis B virus mediated hepatocellular carcinoma. Microb. Pathog. 2019, 128, 184–194. [Google Scholar] [CrossRef]
- Salpini, R.; Surdo, M.; Cortese, M.F.; Palumbo, G.A.; Carioti, L.; Cappiello, G.; Spanò, A.; Trimoulet, P.; Fleury, H.; Vecchiet, J.; et al. The novel HBx mutation F30V correlates with hepatocellular carcinoma in vivo, reduces hepatitis B virus replicative efficiency and enhances anti-apoptotic activity of HBx N terminus in vitro. Clin. Microbiol. Infect. 2019, 25, e901–e906. [Google Scholar] [CrossRef]
- Zhang, T.Y.; Chen, H.Y.; Cao, J.L.; Xiong, H.L.; Mo, X.B.; Li, T.L.; Kang, X.Z.; Zhao, J.H.; Yin, B.; Zhao, X.; et al. Structural and functional analyses of hepatitis B virus X protein BH3-like domain and Bcl-xL interaction. Nat. Commun. 2019, 10, 3192. [Google Scholar] [CrossRef]
- Ma, N.F.; Lau, S.H.; Hu, L.; Xie, D.; Wu, J.; Yang, J.; Wang, Y.; Wu, M.C.; Fung, J.; Bai, X.; et al. COOH-terminal truncated HBV X protein plays key role in hepatocarcinogenesis. Clin. Cancer Res. 2008, 14, 5061–5068. [Google Scholar] [CrossRef]
- Jung, S.Y.; Kim, Y.J. C-terminal region of HBx is crucial for mitochondrial DNA damage. Cancer Lett. 2013, 331, 76–83. [Google Scholar] [CrossRef]
- Gu, C.; Tao, S.; Hu, K.; Ming, L.; Luo, M.; Guo, H.; Su, Y.; Liu, J.; Xie, Y. Establishment of an in vitro reporter system for screening HBx-targeting molecules. Acta Biochim. Biophys. Sin. 2019, 51, 431–440. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, W.; Liu, Q.; Zhang, X.; Lv, N.; Ye, L.; Zhang, X. A mutant of hepatitis B virus X protein (HBxDelta127) promotes cell growth through a positive feedback loop involving 5-lipoxygenase and fatty acid synthase. Neoplasia 2010, 12, 103–115. [Google Scholar] [CrossRef]
- Wang, L.H.; Wu, C.F.; Rajasekaran, N.; Shin, Y.K. Loss of Tumor Suppressor Gene Function in Human Cancer: An Overview. Cell. Physiol. Biochem. 2018, 51, 2647–2693. [Google Scholar] [CrossRef]
- Sartorius, K.; An, P.; Winkler, C.; Chuturgoon, A.; Li, X.; Makarova, J.; Kramvis, A. The Epigenetic Modulation of Cancer and Immune Pathways in Hepatitis B Virus-Associated Hepatocellular Carcinoma: The Influence of HBx and miRNA Dysregulation. Front. Immunol. 2021, 12, 661204. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sun, Y.; Song, X.; Wang, Z.; Zhang, Y.; Zhao, Y.; Peng, X.; Zhang, X.; Li, C.; Gao, C.; et al. Hepatitis B virus evades immune recognition via RNA adenosine deaminase ADAR1-mediated viral RNA editing in hepatocytes. Cell. Mol. Immunol. 2021, 18, 1871–1882. [Google Scholar] [CrossRef] [PubMed]
- Salimi-Jeda, A.; Badrzadeh, F.; Esghaei, M.; Abdoli, A. The role of telomerase and viruses interaction in cancer development, and telomerase-dependent therapeutic approaches. Cancer Treat. Res. Commun. 2021, 27, 100323. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Huang, Y. Antagonism of RIP1 using necrostatin-1 (Nec-1) ameliorated damage and inflammation of HBV X protein (HBx) in human normal hepatocytes. Artif. Cells Nanomed. Biotechnol. 2019, 47, 1194–1199. [Google Scholar] [CrossRef]
- Meirson, T.; Gil-Henn, H.; Samson, A.O. Invasion and metastasis: The elusive hallmark of cancer. Oncogene 2020, 39, 2024–2026. [Google Scholar] [CrossRef]
- You, H.; Yuan, D.; Bi, Y.; Zhang, N.; Li, Q.; Tu, T.; Wei, X.; Lian, Q.; Yu, T.; Kong, D.; et al. Hepatitis B virus X protein promotes vimentin expression via LIM and SH3 domain protein 1 to facilitate epithelial-mesenchymal transition and hepatocarcinogenesis. Cell Commun. Signal. CCS 2021, 19, 33. [Google Scholar] [CrossRef]
- Sanz-Cameno, P.; Martín-Vílchez, S.; Lara-Pezzi, E.; Borque, M.J.; Salmerón, J.; Muñoz de Rueda, P.; Solís, J.A.; López-Cabrera, M.; Moreno-Otero, R. Hepatitis B virus promotes angiopoietin-2 expression in liver tissue: Role of HBV x protein. Am. J. Pathol. 2006, 169, 1215–1222. [Google Scholar] [CrossRef]
- Ahodantin, J.; Bou-Nader, M.; Cordier, C.; Mégret, J.; Soussan, P.; Desdouets, C.; Kremsdorf, D. Hepatitis B virus X protein promotes DNA damage propagation through disruption of liver polyploidization and enhances hepatocellular carcinoma initiation. Oncogene 2019, 38, 2645–2657. [Google Scholar] [CrossRef]
- Bock, C.T.; Toan, N.L.; Koeberlein, B.; Song, L.H.; Chin, R.; Zentgraf, H.; Kandolf, R.; Torresi, J. Subcellular mislocalization of mutant hepatitis B X proteins contributes to modulation of STAT/SOCS signaling in hepatocellular carcinoma. Intervirology 2008, 51, 432–443. [Google Scholar] [CrossRef]
- Lee, A.R.; Lim, K.H.; Park, E.S.; Kim, D.H.; Park, Y.K.; Park, S.; Kim, D.S.; Shin, G.C.; Kang, H.S.; Won, J.; et al. Multiple Functions of Cellular FLIP Are Essential for Replication of Hepatitis B Virus. J. Virol. 2018, 92, e00339-18. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Qi, Y.; Luo, J.; Yang, J.; Xie, Q.; Deng, C.; Su, N.; Wei, W.; Shi, D.; Xu, F.; et al. Hepatitis B Virus X Protein Stimulates Proliferation, Wound Closure and Inhibits Apoptosis of HuH-7 Cells via CDC42. Int. J. Mol. Sci. 2017, 18, 586. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yan, Q.; Gong, L.; Xu, H.; Liu, B.; Fang, X.; Yu, D.; Li, L.; Wei, T.; Wang, Y.; et al. Correction: C-terminal truncated HBx initiates hepatocarcinogenesis by downregulating TXNIP and reprogramming glucose metabolism. Oncogene 2021, 40, 5451–5453. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.Y.; Li, D.; Cai, D.E.; Huang, X.Y.; Zheng, B.Y.; Huang, Y.H.; Chen, Z.X.; Wang, X.Z. Hepatitis B virus X protein sensitizes HL-7702 cells to oxidative stress-induced apoptosis through modulation of the mitochondrial permeability transition pore. Oncol. Rep. 2017, 37, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Kornyeyev, D.; Ramakrishnan, D.; Voitenleitner, C.; Livingston, C.M.; Xing, W.; Hung, M.; Kwon, H.J.; Fletcher, S.P.; Beran, R.K. Spatiotemporal Analysis of Hepatitis B Virus X Protein in Primary Human Hepatocytes. J. Virol. 2019, 93, e00248-19. [Google Scholar] [CrossRef]
- Zhang, J.; Ma, J.; Wang, H.; Guo, L.; Li, J. Serum microRNA-30c levels are correlated with disease progression in Xinjiang Uygur patients with chronic hepatitis B. Braz. J. Med. Biol. Res. 2017, 50, e6050. [Google Scholar] [CrossRef]
- Al-Anazi, M.R.; Nazir, N.; Colak, D.; Al-Ahdal, M.N.; Al-Qahtani, A.A. Deletion and Functional Analysis of Hepatitis B Virus X Protein: Evidence for an Effect on Cell Cycle Regulators. Cell. Physiol. Biochem. 2018, 49, 1987–1998. [Google Scholar] [CrossRef]
- Zha, Y.; Yao, Q.; Liu, J.S.; Wang, Y.Y.; Sun, W.M. Hepatitis B virus X protein promotes epithelial-mesenchymal transition and metastasis in hepatocellular carcinoma cell line HCCLM3 by targeting HMGA2. Oncol. Lett. 2018, 16, 5709–5714. [Google Scholar] [CrossRef]
- Huang, X.Y.; Li, D.; Chen, Z.X.; Huang, Y.H.; Gao, W.Y.; Zheng, B.Y.; Wang, X.Z. Hepatitis B Virus X protein elevates Parkin-mediated mitophagy through Lon Peptidase in starvation. Exp. Cell Res. 2018, 368, 75–83. [Google Scholar] [CrossRef]
- Tse, A.P.; Sze, K.M.; Shea, Q.T.; Chiu, E.Y.; Tsang, F.H.; Chiu, D.K.; Zhang, M.S.; Lee, D.; Xu, I.M.; Chan, C.Y.; et al. Hepatitis transactivator protein X promotes extracellular matrix modification through HIF/LOX pathway in liver cancer. Oncogenesis 2018, 7, 44. [Google Scholar] [CrossRef]
- Duan, L.; Wu, R.; Zhang, X.; Wang, D.; You, Y.; Zhang, Y.; Zhou, L.; Chen, W. HBx-induced S100A9 in NF-κB dependent manner promotes growth and metastasis of hepatocellular carcinoma cells. Cell Death Dis. 2018, 9, 629. [Google Scholar] [CrossRef] [Green Version]
- Kang, H.J.; Chung, D.H.; Sung, C.O.; Yoo, S.H.; Yu, E.; Kim, N.; Lee, S.H.; Song, J.Y.; Kim, C.J.; Choi, J. SHP2 is induced by the HBx-NF-κB pathway and contributes to fibrosis during human early hepatocellular carcinoma development. Oncotarget 2017, 8, 27263–27276. [Google Scholar] [CrossRef] [PubMed]
- Yen, C.J.; Yang, S.T.; Chen, R.Y.; Huang, W.; Chayama, K.; Lee, M.H.; Yang, S.J.; Lai, H.S.; Yen, H.Y.; Hsiao, Y.W.; et al. Hepatitis B virus X protein (HBx) enhances centrosomal P4.1-associated protein (CPAP) expression to promote hepatocarcinogenesis. J. Biomed. Sci. 2019, 26, 44. [Google Scholar] [CrossRef]
- He, Z.; Yu, Y.; Nong, Y.; Du, L.; Liu, C.; Cao, Y.; Bai, L.; Tang, H. Hepatitis B virus X protein promotes hepatocellular carcinoma invasion and metastasis via upregulating thioredoxin interacting protein. Oncol. Lett. 2017, 14, 1323–1332. [Google Scholar] [CrossRef] [PubMed]
- Peantum, J.; Kunanopparat, A.; Hirankarn, N.; Tangkijvanich, P.; Kimkong, I. Autophagy Related-Protein 16-1 Up-Regulated in Hepatitis B Virus-Related Hepatocellular Carcinoma and Impaired Apoptosis. Gastroenterol. Res. 2018, 11, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Laurent, J.M.; Garge, R.K.; Teufel, A.I.; Wilke, C.O.; Kachroo, A.H.; Marcotte, E.M. Humanization of yeast genes with multiple human orthologs reveals functional divergence between paralogs. PLoS Biol. 2020, 18, e3000627. [Google Scholar] [CrossRef] [PubMed]
- Botstein, D.; Fink, G.R. Yeast: An experimental organism for 21st Century biology. Genetics 2011, 189, 695–704. [Google Scholar] [CrossRef]
- Garge, R.K.; Laurent, J.M.; Kachroo, A.H.; Marcotte, E.M. Systematic Humanization of the Yeast Cytoskeleton Discerns Functionally Replaceable from Divergent Human Genes. Genetics 2020, 215, 1153–1169. [Google Scholar] [CrossRef]
- Laurent, J.M.; Young, J.H.; Kachroo, A.H.; Marcotte, E.M. Efforts to make and apply humanized yeast. Brief. Funct. Genom. 2016, 15, 155–163. [Google Scholar] [CrossRef]
- Coronas-Serna, J.M.; Valenti, M.; Del Val, E.; Fernández-Acero, T.; Rodríguez-Escudero, I.; Mingo, J.; Luna, S.; Torices, L.; Pulido, R.; Molina, M.; et al. Modeling human disease in yeast: Recreating the PI3K-PTEN-Akt signaling pathway in Saccharomyces cerevisiae. Int. Microbiol. 2020, 23, 75–87. [Google Scholar] [CrossRef]
- Goleva, T.; Rogov, A.; Zvyagilskaya, R. Alzheimer’s Disease: Molecular Hallmarks and Yeast Models. J. Alzheimer’s Dis. Parkinsonism 2017, 7, 394. [Google Scholar] [CrossRef]
- Sampaio-Marques, B.; Burhans, W.C.; Ludovico, P. Yeast at the Forefront of Research on Ageing and Age-Related Diseases. Prog. Mol. Subcell. Biol. 2019, 58, 217–242. [Google Scholar] [CrossRef] [PubMed]
- Goleva, T.N.; Rogov, A.G.; Korshunova, G.A.; Trendeleva, T.A.; Mamaev, D.V.; Aliverdieva, D.A.; Zvyagilskaya, R.A. SkQThy, a novel and promising mitochondria-targeted antioxidant. Mitochondrion 2019, 49, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Trendeleva, T.; Sukhanova, E.; Ural’skaya, L.; Saris, N.E.; Zvyagilskaya, R. Effect of prooxidants on yeast mitochondria. J. Bioenerg. Biomembr. 2011, 43, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Trendeleva, T.A.; Sukhanova, E.I.; Rogov, A.G.; Zvyagilskaya, R.A.; Seveina, I.I.; Ilyasova, T.M.; Cherepanov, D.A.; Skulachev, V.P. Role of charge screening and delocalization for lipophilic cation permeability of model and mitochondrial membranes. Mitochondrion 2013, 13, 500–506. [Google Scholar] [CrossRef] [PubMed]
- Zvyagilskaya, R.; Parchomenko, O.; Abramova, N.; Allard, P.; Panaretakis, T.; Pattison-Granberg, J.; Persson, B.L. Proton- and sodium-coupled phosphate transport systems and energy status of Yarrowia lipolytica cells grown in acidic and alkaline conditions. J. Membr. Biol. 2001, 183, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Mamaev, D.; Zvyagilskaya, R. Yarrowia lipolytica: A multitalented yeast species of ecological significance. FEMS Yeast Res. 2021, 21, foab008. [Google Scholar] [CrossRef]
- Rogov, A.G.; Ovchenkova, A.P.; Goleva, T.N.; Kireev, I.I.; Zvyagilskaya, R.A. New yeast models for studying mitochondrial morphology as affected by oxidative stress and other factors. Anal. Biochem. 2018, 552, 24–29. [Google Scholar] [CrossRef]
- Tian, Y.; Hu, Y.; Wang, Z.; Chen, K.; Zhang, L.; Wang, L.; Ren, M.; Huang, A.; Tang, H. Hepatitis B virus regulates Raf1 expression in HepG2.2.15 cells by enhancing its promoter activity. Arch. Virol. 2011, 156, 869–874. [Google Scholar] [CrossRef]
- Thakur, S.; Cattoni, D.I.; Nöllmann, M. The fluorescence properties and binding mechanism of SYTOX green, a bright, low photo-damage DNA intercalating agent. Eur. Biophys. J. 2015, 44, 337–348. [Google Scholar] [CrossRef]
- Chance, B.; Williams, G.R. A simple and rapid assay of oxidative phosphorylation. Nature 1955, 175, 1120–1121. [Google Scholar] [CrossRef]
- Akerman, K.E.; Wikström, M.K. Safranine as a probe of the mitochondrial membrane potential. FEBS Lett. 1976, 68, 191–197. [Google Scholar] [CrossRef]
- Zharova, T.V.; Vinogradov, A.D. Energy-linked binding of Pi is required for continuous steady-state proton-translocating ATP hydrolysis catalyzed by F0.F1 ATP synthase. Biochemistry 2006, 45, 14552–14558. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Bertrand, B.; Garduño-Juárez, R.; Munoz-Garay, C. Estimation of pore dimensions in lipid membranes induced by peptides and other biomolecules: A review. Biochim. Biophys. Acta Biomembr. 2021, 1863, 183551. [Google Scholar] [CrossRef] [PubMed]
- Korshunov, S.S.; Skulachev, V.P.; Starkov, A.A. High protonic potential actuates a mechanism of production of reactive oxygen species in mitochondria. FEBS Lett. 1997, 416, 15–18. [Google Scholar] [CrossRef]
- Rogov, A.G.; Goleva, T.N.; Epremyan, K.K.; Kireev, I.I.; Zvyagilskaya, R.A. Propagation of Mitochondria-Derived Reactive Oxygen Species within the Dipodascus magnusii Cells. Antioxidants 2021, 10, 120. [Google Scholar] [CrossRef]
- Chen, H.; Chan, D.C. Emerging functions of mammalian mitochondrial fusion and fission. Hum. Mol. Genet. 2005, 14, R283–R289. [Google Scholar] [CrossRef]
- Simula, L.; Campello, S. Monitoring the Mitochondrial Dynamics in Mammalian Cells. Methods Mol. Biol. 2018, 1782, 267–285. [Google Scholar] [CrossRef]
- Viana, M.P.; Brown, A.I.; Mueller, I.A.; Goul, C.; Koslover, E.F.; Rafelski, S.M. Mitochondrial Fission and Fusion Dynamics Generate Efficient, Robust, and Evenly Distributed Network Topologies in Budding Yeast Cells. Cell Syst. 2020, 10, 287–297.e285. [Google Scholar] [CrossRef]
- Wang, I.H.; Chen, H.Y.; Wang, Y.H.; Chang, K.W.; Chen, Y.C.; Chang, C.R. Resveratrol modulates mitochondria dynamics in replicative senescent yeast cells. PLoS ONE 2014, 9, e104345. [Google Scholar] [CrossRef]
- Millare, B.; O’Rourke, B.; Trayanova, N. Hydrogen peroxide diffusion and scavenging shapes mitochondrial network instability and failure by sensitizing ROS-induced ROS release. Sci. Rep. 2020, 10, 15758. [Google Scholar] [CrossRef] [PubMed]
- Zorov, D.B.; Juhaszova, M.; Sollott, S.J. Mitochondrial ROS-induced ROS release: An update and review. Biochim. Biophys. Acta 2006, 1757, 509–517. [Google Scholar] [CrossRef]
- Feniouk, B.A.; Skulachev, V.P. Cellular and Molecular Mechanisms of Action of Mitochondria-Targeted Antioxidants. Curr. Aging Sci. 2017, 10, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Baek, S.H.; Park, S.J.; Jeong, J.I.; Kim, S.H.; Han, J.; Kyung, J.W.; Baik, S.H.; Choi, Y.; Choi, B.Y.; Park, J.S.; et al. Inhibition of Drp1 Ameliorates Synaptic Depression, Aβ Deposition, and Cognitive Impairment in an Alzheimer’s Disease Model. J. Neurosci. 2017, 37, 5099–5110. [Google Scholar] [CrossRef] [PubMed]
- Lefranc, C.; Friederich-Persson, M.; Palacios-Ramirez, R.; Nguyen Dinh Cat, A. Mitochondrial oxidative stress in obesity: Role of the mineralocorticoid receptor. J. Endocrinol. 2018, 238, R143–R159. [Google Scholar] [CrossRef]
- Ong, S.B.; Kwek, X.Y.; Katwadi, K.; Hernandez-Resendiz, S.; Crespo-Avilan, G.E.; Ismail, N.I.; Lin, Y.H.; Yap, E.P.; Lim, S.Y.; Ja, K.; et al. Targeting Mitochondrial Fission Using Mdivi-1 in A Clinically Relevant Large Animal Model of Acute Myocardial Infarction: A Pilot Study. Int. J. Mol. Sci. 2019, 20, 3972. [Google Scholar] [CrossRef]
- Reddy, P.H.; Manczak, M.; Yin, X. Mitochondria-Division Inhibitor 1 Protects Against Amyloid-β induced Mitochondrial Fragmentation and Synaptic Damage in Alzheimer’s Disease. J. Alzheimer’s Dis. JAD 2017, 58, 147–162. [Google Scholar] [CrossRef] [Green Version]










| Strain | Description |
|---|---|
| Po1f | MatA, leu2-270, ura3-302, xpr2-322, axp-2 |
| pZ-0 | Po1f + pZ URA3, xpr2Δ |
| pZ-eGFP | Po1f + pZ-eGFP URA3 xpr2Δ |
| pZ-HBx | Po1f + pZ-HBx URA3 xpr2Δ |
| pZ-HBx-eGFP | Po1f + pZ-HBx-eGFP URA3 xpr2Δ |
| Primer | Sequence |
|---|---|
| HBx-BbsI-Fw1 | TAGAAGACGCAATGGCTGCTAGGCTGTGC |
| HBx-BbsI-Rev1 | TAGAAGACGCGCGCTTAGGCAGAGGTGAAAAAGTTGC |
| HBx-BbsI-rev2 | TAGAAGACGCGGCAGAGGTGAAAAAGTTGC |
| eGFP-BbsI-Fw1 | TAGAAGACTAAATGGTGAGCAAGGGCGAGGAG |
| eGFP-BbsI-Rev1 | TAGAAGACGCGCGCTTACTTGTACAGCTCGTCCATG |
| eGFP-BbsI-Fw2 | TAGAAGACATTGCCATGGTGAGCAAGGGCGAG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Epremyan, K.K.; Goleva, T.N.; Rogov, A.G.; Lavrushkina, S.V.; Zinovkin, R.A.; Zvyagilskaya, R.A. The First Yarrowia lipolytica Yeast Models Expressing Hepatitis B Virus X Protein: Changes in Mitochondrial Morphology and Functions. Microorganisms 2022, 10, 1817. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms10091817
Epremyan KK, Goleva TN, Rogov AG, Lavrushkina SV, Zinovkin RA, Zvyagilskaya RA. The First Yarrowia lipolytica Yeast Models Expressing Hepatitis B Virus X Protein: Changes in Mitochondrial Morphology and Functions. Microorganisms. 2022; 10(9):1817. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms10091817
Chicago/Turabian StyleEpremyan, Khoren K., Tatyana N. Goleva, Anton G. Rogov, Svetlana V. Lavrushkina, Roman A. Zinovkin, and Renata A. Zvyagilskaya. 2022. "The First Yarrowia lipolytica Yeast Models Expressing Hepatitis B Virus X Protein: Changes in Mitochondrial Morphology and Functions" Microorganisms 10, no. 9: 1817. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms10091817