Arbuscular Mycorrhizal Fungal Assemblages Significantly Shifted upon Bacterial Inoculation in Non-Contaminated and Petroleum-Contaminated Environments
Abstract
:1. Introduction
2. Materials and Methods
2.1. Seeds Collection and Germination
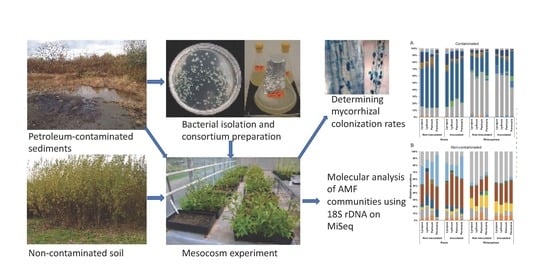
2.2. Media Preparation and Isolation of Bacteria from the Contaminated Sediments
2.3. 16S rDNA Amplification, Sequencing, and Identification of Bacteria
2.4. Selection of Isolates and Production of a Bacterial Consortium
2.5. Mesocosm Experiment Design and Inoculation
2.6. Data Collection and Harvest
2.7. DNA Extraction, PCR Amplification and Illumina MiSeq Sequencing
2.8. Evaluation of the AMF Colonization of Plants Roots
2.9. Sequence Processing and Details of the Pipeline
2.10. Statistical Analyses
2.11. Phylogenetic Analysis
3. Results
3.1. Sequence Processing
3.2. Diversity and Identity of AMF Taxa
3.3. AMF Community Structure
3.4. Plant Dry Biomass
3.5. AMF Root Colonization
3.6. Effect of Inoculation on PH Concentrations
4. Discussion
4.1. Contamination and Biotope Shape AMF Communities
4.2. Glomeraceae and Claroideoglomeraceae Dominate Most Samples
4.3. Contamination Increases Root AM Colonization
4.4. Inoculation Affects Plant Growth and PH Attenuation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Van Der Heijden, M.G.A.; Bardgett, R.D.; Van Straalen, N.M. The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 2008, 11, 296–310. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.; Bakker, P.A. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Ping, L.; Boland, W. Signals from the underground: Bacterial volatiles promote growth in Arabidopsis. Trends Plant Sci. 2004, 9, 263–266. [Google Scholar] [CrossRef]
- Dangl, J.L.; Horvath, D.M.; Staskawicz, B.J. Pivoting the Plant Immune System from Dissection to Deployment. Science 2013, 341, 746–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jambon, I.; Thijs, S.; Weyens, N.; Vangronsveld, J. Harnessing plant-bacteria-fungi interactions to improve plant growth and degradation of organic pollutants. J. Plant Interact. 2018, 13, 119–130. [Google Scholar] [CrossRef] [Green Version]
- Ojuederie, O.B.; Babalola, O.O. Microbial and Plant-Assisted Bioremediation of Heavy Metal Polluted Environments: A Review. Int. J. Environ. Res. Public Health 2017, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gkorezis, P.; Daghio, M.; Franzetti, A.; Van Hamme, J.D.; Sillen, W.; Vangronsveld, J. The Interaction between Plants and Bacteria in the Remediation of Petroleum Hydrocarbons: An Environmental Perspective. Front. Microbiol. 2016, 7, 1836. [Google Scholar] [CrossRef] [PubMed]
- Alori, E.T.; Dare, M.O.; Babalola, O.O. Microbial Inoculants for Soil Quality and Plant Health. In Sustainable Agriculture Reviews; Lichtfouse, E., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 281–307. [Google Scholar] [CrossRef]
- Fox, J.L. Agricultural probiotics enter spotlight. Nat. Publ. 2015, 33, 122. [Google Scholar] [CrossRef] [PubMed]
- Kaminsky, L.M.; Trexler, R.V.; Malik, R.J.; Hockett, K.L.; Bell, T.H. The Inherent Conflicts in Developing Soil Microbial Inoculants. Trends Biotechnol. 2019, 37, 140–151. [Google Scholar] [CrossRef]
- Ratzke, C.; Gore, J. Modifying and reacting to the environmental pH can drive bacterial interactions. PLoS Biol. 2018, 16, e2004248. [Google Scholar] [CrossRef] [Green Version]
- Kallala, N.; M’sehli, W.; Jelali, K.; Kais, Z.; Mhadhbi, H. Inoculation with Efficient Nitrogen Fixing and Indoleacetic Acid Producing Bacterial Microsymbiont Enhance Tolerance of the Model Legume Medicago truncatula to Iron Deficiency. BioMed Res. Int. 2018, 2018, 14. [Google Scholar] [CrossRef] [Green Version]
- Yergeau, E.; Bell, T.; Champagne, J.; Maynard, C.; Tardif, S.; Tremblay, J.; Greer, C. Transplanting soil microbiomes leads to lasting effects on willow growth, but not on the rhizosphere microbiome. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Dagher, D.J.; de la Providencia, I.E.; Pitre, F.E.; St-Arnaud, M.; Hijri, M. Plant Identity Shaped Rhizospheric Microbial Communities More Strongly Than Bacterial Bioaugmentation in Petroleum Hydrocarbon-Polluted Sediments. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Smith, S.E.; Read, D.J. Mycorrhizal Symbiosis; Elsevier: Amsterdam, The Netherlands, 2010; Available online: ScienceDirect.com (accessed on 17 January 2020).
- Abdel Latef, A.A.H.; Chaoxing, H. Effect of arbuscular mycorrhizal fungi on growth, mineral nutrition, antioxidant enzymes activity and fruit yield of tomato grown under salinity stress. Sci. Hortic. 2011, 127, 228–233. [Google Scholar] [CrossRef]
- Jakobsen, I.; Smith, S.E.; Smith, F.A. Function and Diversity of Arbuscular Mycorrhizae in Carbon and Mineral Nutrition. In Mycorrhizal Ecology; van der Heijden, M.G.A., Sanders, I.R., Eds.; Springer Berlin Heidelberg: Berlin/Heidelberg, Germany, 2003. [Google Scholar]
- Posta, K.; Duc, N.H. Benefits of Arbuscular Mycorrhizal Fungi Application to Crop Production under Water Scarcity. In Drought (Aridity); IntechOpen: London, UK, 2019. [Google Scholar]
- Wu, Z.; McGrouther, K.; Huang, J.; Wu, P.; Wu, W.; Wang, H. Decomposition and the contribution of glomalin-related soil protein (GRSP) in heavy metal sequestration: Field experiment. Soil Biol. Biochem. 2014, 68, 283–290. [Google Scholar] [CrossRef]
- Hajiboland, R.; Aliasgharzadeh, N.; Laiegh, S.F.; Poschenrieder, C. Colonization with arbuscular mycorrhizal fungi improves salinity tolerance of tomato (Solanum lycopersicum L.) plants. Plant and Soil 2010, 331, 313–327. [Google Scholar] [CrossRef]
- Begum, N.; Qin, C.; Ahanger, M.A.; Raza, S.; Khan, M.I.; Ashraf, M.; Ahmed, N.; Zhang, L. Role of Arbuscular Mycorrhizal Fungi in Plant Growth Regulation: Implications in Abiotic Stress Tolerance. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; Chen, D.; Lu, K.; Sun, Z.; Zeng, R. Enhanced tomato disease resistance primed by arbuscular mycorrhizal fungus. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Bruisson, S.; Maillot, P.; Schellenbaum, P.; Walter, B.; Gindro, K.; Deglène-Benbrahim, L. Arbuscular mycorrhizal symbiosis stimulates key genes of the phenylpropanoid biosynthesis and stilbenoid production in grapevine leaves in response to downy mildew and grey mould infection. Phytochemistry 2016, 131, 92–99. [Google Scholar] [CrossRef]
- St-Arnaud, M.; Vujanovic, V. Effect of the arbuscular mycorrhizal symbiosis on plant diseases and pests. In Mycorrhizae in Crop Production; Haworth: New York, NY, USA, 2007; pp. 67–122. [Google Scholar]
- Rillig, M.C.; Mummey, D.L. Mycorrhizas and soil structure. New Phytol. 2006, 171, 41–53. [Google Scholar] [CrossRef]
- Rillig, M.C.; Wright, S.F.; Eviner, V.T. The role of arbuscular mycorrhizal fungi and glomalin in soil aggregation: Comparing effects of five plant species. Plant and Soil 2002, 238, 325–333. [Google Scholar] [CrossRef]
- Bedini, S.; Pellegrino, E.; Avio, L.; Pellegrini, S.; Bazzoffi, P.; Argese, E.; Giovannetti, M. Changes in soil aggregation and glomalin-related soil protein content as affected by the arbuscular mycorrhizal fungal species Glomus mosseae and Glomus intraradices. Soil Biol. Biochem. 2009, 41, 1491–1496. [Google Scholar] [CrossRef]
- Joner, E.J.; Johansen, A.; Loibner, A.P.; dela Cruz, M.A.; Szolar, O.H.J.; Portal, J.-M.; Leyval, C. Rhizosphere Effects on Microbial Community Structure and Dissipation and Toxicity of Polycyclic Aromatic Hydrocarbons (PAHs) in Spiked Soil. Environ. Sci. Technol. 2001, 35, 2773–2777. [Google Scholar] [CrossRef]
- Joner, E.; Leyval, C. Phytoremediation of organic pollutants using mycorrhizal plants: A new aspect of rhizosphere interactions. In Sustainable Agriculture; Springer: Dordrecht, The Netherlands, 2003; Volume 23, pp. 495–502. [Google Scholar]
- Barea, J.; Gryndler, M.; Lemanceau, P.; Schüepp, H.; Azcón, R. The rhizosphere of mycorrhizal plants. In Mycorrhizal Technology in Agriculture; Springer: New York, NY, USA, 2002; pp. 1–18. [Google Scholar]
- Jeffries, P.; Gianinazzi, S.; Perotto, S.; Turnau, K.; Barea, J.-M. The contribution of arbuscular mycorrhizal fungi in sustainable maintenance of plant health and soil fertility. Biol. Fertility Soils 2003, 37, 1–16. [Google Scholar] [CrossRef]
- Li, Q.; Ling, W.; Gao, Y.; Li, F.; Xiong, W. Arbuscular mycorrhizal bioremediation and its mechanisms of organic pollutants-contaminated soils. Ying yong sheng tai xue bao = J. Appl. Ecol. 2006, 17, 2217–2221. [Google Scholar]
- Lecomte, J.; St-Arnaud, M.; Hijri, M. Isolation and identification of soil bacteria growing at the expense of arbuscular mycorrhizal fungi. FEMS Microbiol. Lett. 2011, 317, 43–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheublin, T.R.; Sanders, I.R.; Keel, C.; van der Meer, J.R. Characterisation of microbial communities colonising the hyphal surfaces of arbuscular mycorrhizal fungi. ISME J. 2010, 4, 752–763. [Google Scholar] [CrossRef] [Green Version]
- Taktek, S.; St-Arnaud, M.; Piché, Y.; Fortin, J.A.; Antoun, H. Igneous phosphate rock solubilization by biofilm-forming mycorrhizobacteria and hyphobacteria associated with Rhizoglomus irregulare DAOM 197198. Mycorrhiza 2017, 27, 13–22. [Google Scholar] [CrossRef] [Green Version]
- Emam, T. Local soil, but not commercial AMF inoculum, increases native and non-native grass growth at a mine restoration site. Restor. Ecol. 2016, 24, 35–44. [Google Scholar] [CrossRef]
- Desjardins, D.; Nissim, W.G.; Pitre, F.E.; Naud, A.; Labrecque, M. Distribution patterns of spontaneous vegetation and pollution at a former decantation basin in southern Québec, Canada. Ecol. Eng. 2014, 64, 385–390. [Google Scholar] [CrossRef]
- Bell, T.H.; Hassan, S.E.-D.; Lauron-Moreau, A.; Al-Otaibi, F.; Hijri, M.; Yergeau, E.; St-Arnaud, M. Linkage between bacterial and fungal rhizosphere communities in hydrocarbon-contaminated soils is related to plant phylogeny. ISME J. 2014, 8, 331–343. [Google Scholar] [CrossRef] [Green Version]
- Stefani, F.O.; Bell, T.H.; Marchand, C.; de la Providencia, I.E.; El Yassimi, A.; St-Arnaud, M.; Hijri, M. Culture-dependent and-independent methods capture different microbial community fractions in hydrocarbon-contaminated soils. PLoS ONE 2015, 10, e0128272. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Lee, S.; Young, J.P. Improved PCR primers for the detection and identification of arbuscular mycorrhizal fungi. FEMS Microbiol. Ecol. 2008, 65, 339–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renaut, S.; Daoud, R.; Masse, J.; Vialle, A.; Hijri, M. Inoculation with Rhizophagus Irregularis Does Not Alter Arbuscular Mycorrhizal Fungal Community Structure within the Roots of Corn, Wheat, and Soybean Crops. Microorganisms 2020, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalpé, Y.; Séguin, S.M. Microwave-assisted technology for the clearing and staining of arbuscular mycorrhizal fungi in roots. Mycorrhiza 2013, 23, 333–340. [Google Scholar] [CrossRef] [PubMed]
- McGonigle, T.P.; Miller, M.H.; Evans, D.G.; Fairchild, G.L.; Swan, J.A. A new method which gives an objective measure of colonization of roots by vesicular—arbuscular mycorrhizal fungi. New Phytol. 1990, 115, 495–501. [Google Scholar] [CrossRef]
- Pylro, V.S.; Roesch, L.F.W.; Morais, D.K.; Clark, I.M.; Hirsch, P.R.; Tótola, M.R. Data analysis for 16S microbial profiling from different benchtop sequencing platforms. J. Microbiol. Methods 2014, 107, 30–37. [Google Scholar] [CrossRef] [Green Version]
- Pylro, V.S.; Roesch, L.F.W.; Ortega, J.M.; Amaral, A.M.; Tótola, M.R.; Hirsch, P.R.; Rosado, A.S.; Góes-Neto, A.; da Costa da Silva, A.L.; Rosa, C.A.; et al. Brazilian Microbiome Project: Revealing the Unexplored Microbial Diversity—Challenges and Prospects. Microb. Ecol. 2013, 67, 237–241. [Google Scholar] [CrossRef] [Green Version]
- Kruger, M.; Kruger, C.; Walker, C.; Stockinger, H.; Schussler, A. Phylogenetic reference data for systematics and phylotaxonomy of arbuscular mycorrhizal fungi from phylum to species level. New Phytol. 2012, 193, 970–984. [Google Scholar] [CrossRef]
- Öpik, M.; Vanatoa, A.; Vanatoa, E.; Moora, M.; Davison, J.; Kalwij, J.M.; Reier, Ü.; Zobel, M. The online database MaarjAM reveals global and ecosystemic distribution patterns in arbuscular mycorrhizal fungi (Glomeromycota). New Phytol. 2010, 188, 223–241. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics (Oxf., Engl. ) 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [Green Version]
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronquist, F.; Huelsenbeck, J.P. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics (Oxf., Engl.) 2001, 17, 754–755. [Google Scholar] [CrossRef] [Green Version]
- Pielou, E.C. The measurement of diversity in different types of biological collections. J. Theor. Biol. 1966, 13, 131–144. [Google Scholar] [CrossRef]
- Xu, X.; Chen, C.; Zhang, Z.; Sun, Z.; Chen, Y.; Jiang, J.; Shen, Z. The influence of environmental factors on communities of arbuscular mycorrhizal fungi associated with Chenopodium ambrosioides revealed by MiSeq sequencing investigation. Sci. Rep. 2017, 7, 45134. [Google Scholar] [CrossRef]
- Krüger, C.; Kohout, P.; Janoušková, M.; Püschel, D.; Frouz, J.; Rydlová, J. Plant Communities Rather than Soil Properties Structure Arbuscular Mycorrhizal Fungal Communities along Primary Succession on a Mine Spoil. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Wang, F.; Che, R.; Wang, P.; Liu, H.; Ji, B.; Cui, X. Precipitation shapes communities of arbuscular mycorrhizal fungi in Tibetan alpine steppe. Sci. Rep. 2016, 6, 23488. [Google Scholar] [CrossRef]
- Dumbrell, A.J.; Nelson, M.; Helgason, T.; Dytham, C.; Fitter, A.H. Relative roles of niche and neutral processes in structuring a soil microbial community. ISME J. 2009, 4, 337. [Google Scholar] [CrossRef] [Green Version]
- Tropini, C.; Moss, E.L.; Merrill, B.D.; Ng, K.M.; Higginbottom, S.K.; Casavant, E.P.; Gonzalez, C.G.; Fremin, B.; Bouley, D.M.; Elias, J.E.; et al. Transient Osmotic Perturbation Causes Long-Term Alteration to the Gut Microbiota. Cell 2018, 173, 1742–1754. [Google Scholar] [CrossRef] [Green Version]
- Iffis, B.; St-Arnaud, M.; Hijri, M. Petroleum Contamination and Plant Identity Influence Soil and Root Microbial Communities While AMF Spores Retrieved from the Same Plants Possess Markedly Different Communities. Front. Plant Sci. 2017, 8, 1381. [Google Scholar] [CrossRef]
- Iffis, B.; St-Arnaud, M.; Hijri, M. Petroleum hydrocarbon contamination, plant identity and arbuscular mycorrhizal fungal (AMF) community determine assemblages of the AMF spore-associated microbes. Environ. Microbiol. 2016, 18, 2689–2704. [Google Scholar] [CrossRef]
- Taktek, S.; Trépanier, M.; Servin, P.M.; St-Arnaud, M.; Piché, Y.; Fortin, J.A.; Antoun, H. Trapping of phosphate solubilizing bacteria on hyphae of the arbuscular mycorrhizal fungus Rhizophagus irregularis DAOM 197198. Soil Biol. Biochem. 2015, 90, 1–9. [Google Scholar] [CrossRef]
- Miransari, M. Interactions between arbuscular mycorrhizal fungi and soil bacteria. Appl. Microbiol. Biotechnol. 2011, 89, 917–930. [Google Scholar] [CrossRef]
- De la Providencia, I.E.; Stefani, F.O.P.; Labridy, M.; St-Arnaud, M.; Hijri, M. Arbuscular mycorrhizal fungal diversity associated with Eleocharis obtusa and Panicum capillare growing in an extreme petroleum hydrocarbon-polluted sedimentation basin. FEMS Microbiol. Lett. 2015, 362. [Google Scholar] [CrossRef] [Green Version]
- Hassan, S.E.-D.; Bell, T.H.; Stefani, F.O.P.; Denis, D.; Hijri, M.; St-Arnaud, M. Contrasting the Community Structure of Arbuscular Mycorrhizal Fungi from Hydrocarbon-Contaminated and Uncontaminated Soils following Willow (Salix spp. L.) Planting. PLoS ONE 2014, 9, e102838. [Google Scholar] [CrossRef] [Green Version]
- Hart, M.M.; Reader, R.J. The role of the external mycelium in early colonization for three arbuscular mycorrhizal fungal species with different colonization strategies. Pedobiologia 2005, 49, 269–279. [Google Scholar] [CrossRef]
- Hart, M.M.; Reader, R.J. Taxonomic basis for variation in the colonization strategy of arbuscular mycorrhizal fungi. New Phytol. 2002, 153, 335–344. [Google Scholar] [CrossRef]
- Cabello, M.N. Hydrocarbon pollution: Its effect on native arbuscular mycorrhizal fungi (AMF). FEMS Microbiol. Ecol. 1997, 22, 233–236. [Google Scholar] [CrossRef]
- Garcés-Ruiz, M.; Senés-Guerrero, C.; Declerck, S.; Cranenbrouck, S. Arbuscular Mycorrhizal Fungal Community Composition in Carludovica palmata, Costus scaber and Euterpe precatoria from Weathered Oil Ponds in the Ecuadorian Amazon. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Varela-Cervero, S.; López-García, Á.; Barea, J.M.; Azcón-Aguilar, C. Differences in the composition of arbuscular mycorrhizal fungal communities promoted by different propagule forms from a Mediterranean shrubland. Mycorrhiza 2016, 26, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Tuomi, J.; Niemelä, P.; Haukioja, E.; Sirén, S.; Neuvonen, S. Nutrient stress: An explanation for plant anti-herbivore responses to defoliation. Oecologia 1984, 61, 208–210. [Google Scholar] [CrossRef] [PubMed]
- IJdo, M.; Schtickzelle, N.; Cranenbrouck, S.; Declerck, S. Do arbuscular mycorrhizal fungi with contrasting life-history strategies differ in their responses to repeated defoliation? FEMS Microbiol. Ecol. 2010, 72, 114–122. [Google Scholar] [CrossRef]
- Piippo, S.; Markkola, A.; Härmä, E.; Tuomi, J. Do compensatory shoot growth and mycorrhizal symbionts act as competing above- and below-ground sinks after simulated grazing? Plant Ecol. 2011, 212, 33–42. [Google Scholar] [CrossRef]
- Saito, K.; Suyama, Y.; Sato, S.; Sugawara, K. Defoliation effects on the community structure of arbuscular mycorrhizal fungi based on 18S rDNA sequences. Mycorrhiza 2004, 14, 363–373. [Google Scholar] [CrossRef]
- Ambrosino, M.L.; Busso, C.A.; Cabello, M.N.; Velázquez, M.S.; Torres, Y.A.; Ithurrart, L.S.; Cardillo, D.S.; Palomo, I.R. Total and structure colonization by arbuscular mycorrhizal fungi in native, perennial grasses of different forage quality exposed to defoliation. J. King Saud Univ. Sci. 2018, 32, 377–383. [Google Scholar] [CrossRef]
- Svenningsen, N.B.; Watts-Williams, S.J.; Joner, E.J.; Battini, F.; Efthymiou, A.; Cruz-Paredes, C.; Nybroe, O.; Jakobsen, I. Suppression of the activity of arbuscular mycorrhizal fungi by the soil microbiota. ISME J. 2018, 12, 1296–1307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, F.; Yu, X.; Wu, S.; Wong, M. Effects of Inoculation of PAH-degrading Bacteria and Arbuscular Mycorrhizal Fungi on Responses of Ryegrass to Phenanthrene and Pyrene. Int. J. Phytoremediation 2014, 16, 109–122. [Google Scholar] [CrossRef] [PubMed]
- Bell, T.H.; Stefani, F.O.P.; Abram, K.; Champagne, J.; Yergeau, E.; Hijri, M.; St-Arnaud, M. A Diverse Soil Microbiome Degrades More Crude Oil than Specialized Bacterial Assemblages Obtained in Culture. Appl. Environ. Microbiol. 2016, 82, 5530–5541. [Google Scholar] [CrossRef] [Green Version]





| Shannon | Chao 1 | Pielou’s Equitability | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Contamination | Inoculation | Biotope | Plant Species | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev |
| Contaminated | Inoculated | Rhizosphere | LE | 1.7 | 0.38 | 9.96 | 3.35 | 0.56 | 0.12 |
| LS | 1.96 | 0.43 | 8.92 | 2.37 | 0.64 | 0.14 | |||
| PC | 1.94 | 0.43 | 10.88 | 4.43 | 0.62 | 0.13 | |||
| PL | 1.61 | 0.39 | 8.6 | 3.57 | 0.57 | 0.13 | |||
| Roots | LE | 2.39 | 0.22 | 10.87 | 1.38 | 0.71 | 0.07 | ||
| LS | 1.87 | 0.64 | 10.32 | 3.67 | 0.57 | 0.16 | |||
| PC | 2.08 | 0.27 | 8.58 | 1.43 | 0.69 | 0.07 | |||
| PL | 1.73 | 0.5 | 8.42 | 3.05 | 0.58 | 0.11 | |||
| Non-inoculated | Rhizosphere | LE | 1.86 | 0.36 | 9.3 | 2.82 | 0.62 | 0.11 | |
| LS | 1.87 | 0.5 | 7.74 | 2.16 | 0.65 | 0.14 | |||
| PC | 1.74 | 0.65 | 8.86 | 4.5 | 0.61 | 0.14 | |||
| PL | 1.68 | 0.32 | 7.7 | 2.63 | 0.62 | 0.12 | |||
| Roots | LE | 2.09 | 0.27 | 9.25 | 1.78 | 0.67 | 0.07 | ||
| LS | 2.25 | 0.42 | 17.5 | 2.93 | 0.6 | 0.09 | |||
| PC | 1.75 | 0.62 | 8.58 | 2.85 | 0.59 | 0.15 | |||
| PL | 1.39 | 0.51 | 8.36 | 3.71 | 0.48 | 0.13 | |||
| Non-contaminated | Inoculated | Rhizosphere | LE | 2.51 | 0.28 | 13.52 | 4.42 | 0.71 | 0.07 |
| LS | 2.49 | 0.25 | 12 | 2.5 | 0.71 | 0.04 | |||
| PC | 2.6 | 0.34 | 12.68 | 2.08 | 0.73 | 0.07 | |||
| PL | 2.79 | 0.31 | 15.3 | 5.96 | 0.76 | 0.06 | |||
| Roots | LE | 2.92 | 0.67 | 14.94 | 2.87 | 0.76 | 0.15 | ||
| LS | 3.05 | 0.22 | 14.43 | 1.65 | 0.8 | 0.06 | |||
| PC | 2.82 | 0.34 | 14.24 | 3.34 | 0.77 | 0.08 | |||
| PL | 2.79 | 0.52 | 13.49 | 1.95 | 0.75 | 0.12 | |||
| Non-inoculated | Rhizosphere | LE | 2.33 | 0.37 | 14.87 | 4.16 | 0.65 | 0.09 | |
| LS | 2.43 | 0.33 | 12.27 | 1.69 | 0.68 | 0.08 | |||
| PC | 2.28 | 0.44 | 10.69 | 2.55 | 0.68 | 0.09 | |||
| PL | 2.46 | 0.57 | 14.24 | 3.1 | 0.67 | 0.12 | |||
| Roots | LE | 2.7 | 0.93 | 14 | 5.29 | 0.73 | 0.23 | ||
| LS | 2.82 | 0.46 | 14.18 | 2.82 | 0.77 | 0.11 | |||
| PC | 2.59 | 0.47 | 12.95 | 2.43 | 0.73 | 0.11 | |||
| PL | 2.47 | 0.31 | 13.06 | 1.85 | 0.69 | 0.07 | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dagher, D.J.; de la Providencia, I.E.; Pitre, F.E.; St-Arnaud, M.; Hijri, M. Arbuscular Mycorrhizal Fungal Assemblages Significantly Shifted upon Bacterial Inoculation in Non-Contaminated and Petroleum-Contaminated Environments. Microorganisms 2020, 8, 602. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms8040602
Dagher DJ, de la Providencia IE, Pitre FE, St-Arnaud M, Hijri M. Arbuscular Mycorrhizal Fungal Assemblages Significantly Shifted upon Bacterial Inoculation in Non-Contaminated and Petroleum-Contaminated Environments. Microorganisms. 2020; 8(4):602. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms8040602
Chicago/Turabian StyleDagher, Dimitri J., Ivan E. de la Providencia, Frédéric E. Pitre, Marc St-Arnaud, and Mohamed Hijri. 2020. "Arbuscular Mycorrhizal Fungal Assemblages Significantly Shifted upon Bacterial Inoculation in Non-Contaminated and Petroleum-Contaminated Environments" Microorganisms 8, no. 4: 602. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms8040602