Unraveling Mechanisms and Impact of Microbial Recruitment on Oilseed Rape (Brassica napus L.) and the Rhizosphere Mediated by Plant Growth-Promoting Rhizobacteria
Abstract
:1. Introduction
2. Materials and Methods
2.1. PGPR Inoculants and Plants
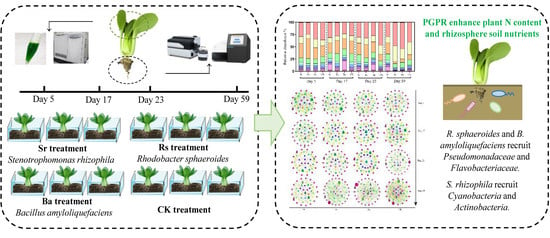
2.2. Greenhouse Pot Experiment
2.3. Plant Physicochemical Parameters
2.4. Rhizosphere and Bulk Soil Collection and Physicochemical Parameters
2.5. Evaluation of Plant Growth Promotion Characteristics
2.6. DNA Extraction and 16S rRNA Sequencing
2.7. The Quantification PCR of the PGPR and N Cycling Functional Genes in the Rhizosphere
2.8. Network Construction with RMT-Based Approach and Topological Analysis
2.9. Data Analysis
3. Results
3.1. PGPR Promote Plant Growth and Enhance the Nutrient Availability in the Rhizosphere
3.2. Identification of PGPR Promoting Abilities
3.3. Low, Persistent Colonization of PGPR Strains in the Rhizosphere
3.4. S. rhizophila Increased the Ammonification in the Rhizosphere
3.5. Rhizobacterial Community Assembly Is Driven by Plant Growth Stage and PGPR
3.6. Dynamic Rhizobacteria Interactions during Plant Growth
4. Discussion
4.1. Plant Growth Stages Determine Rhizobacterial Community Composition
4.2. R. sphaeroides and B. amyloliquefaciens Increase the Selective Enrichment of Beneficial Bacteria in the Plant Rhizosphere
4.3. The Difference of S. rhizophila Application to the Rhizobacterial Community
4.4. PGPR Promote Plant Growth and Enhance Soil Nutrient Availability by Shaping the Rhizobacterial Community
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Venturi, V.; Keel, C. Signaling in the Rhizosphere. Trends Plant Sci. 2016, 21, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Verbon, E.H.; Liberman, L.M. Beneficial Microbes Affect Endogenous Mechanisms Controlling Root Development. Trends Plant Sci. 2016, 21, 218–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malik, K.; Bilal, R.; Mehnaz, S.; Rasul, G.; Mirza, M.; Ali, S. Association of nitrogen-fixing, plant-growth-promoting rhizobacteria (PGPR) with kallar grass and rice. In Opportunities for Biological Nitrogen Fixation in Rice and Other Non-Legumes; Springer: Cham, Switzerland, 1997; pp. 37–44. [Google Scholar]
- Jog, R.; Pandya, M.; Nareshkumar, G.; Rajkumar, S. Mechanism of phosphate solubilization and antifungal activity of Streptomyces spp. isolated from wheat roots and rhizosphere and their application in improving plant growth. Microbiology 2014, 160, 778–788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kloepper, J.W.; Leong, J.; Teintze, M.; Schroth, M.N. Enhanced plant growth by siderophores produced by plant growth-promoting rhizobacteria. Nature 1980, 286, 885–886. [Google Scholar] [CrossRef]
- Bal, H.B.; Nayak, L.; Das, S.; Adhya, T.K. Isolation of ACC deaminase producing PGPR from rice rhizosphere and evaluating their plant growth promoting activity under salt stress. Plant Soil 2013, 366, 93–105. [Google Scholar] [CrossRef]
- Cassán, F.; Vanderleyden, J.; Spaepen, S. Physiological and agronomical aspects of phytohormone production by model plant-growth-promoting rhizobacteria (PGPR) belonging to the genus Azospirillum. J. Plant Growth Regul. 2014, 33, 440–459. [Google Scholar] [CrossRef]
- Ramamoorthy, V.; Viswanathan, R.; Raguchander, T.; Prakasam, V.; Samiyappan, R. Induction of systemic resistance by plant growth promoting rhizobacteria in crop plants against pests and diseases. Crop Prot. 2001, 20, 1–11. [Google Scholar] [CrossRef]
- Bhattacharyya, P.N.; Jha, D.K. Plant growth-promoting rhizobacteria (PGPR): Emergence in agriculture. World J. Microbiol. Biotechnol. 2012, 28, 1327–1350. [Google Scholar] [CrossRef]
- Wu, S.; Zhuang, G.; Bai, Z.; Cen, Y.; Xu, S.; Sun, H.; Han, X.; Zhuang, X. Mitigation of nitrous oxide emissions from acidic soils by Bacillus amyloliquefaciens, a plant growth-promoting bacterium. Glob. Chang. Biol. 2018, 24, 2352–2365. [Google Scholar] [CrossRef]
- Deng, Y.; Liu, X.; Wu, J.; Lee, J.; Chen, S.; Cheng, Y.; Zhang, C.; Zhang, L.H. The host plant metabolite glucose is the precursor of diffusible signal factor (DSF) family signals in Xanthomonas campestris. Appl. Environ. Microbiol. 2015, 81, 2861–2868. [Google Scholar] [CrossRef] [Green Version]
- Tan, S.Y.; Gu, Y.; Yang, C.L.; Dong, Y.; Mei, X.L.; Shen, Q.R.; Xu, Y.C. Bacillus amyloliquefaciens T-5 may prevent Ralstonia solanacearum infection through competitive exclusion. Biol. Fertil. Soils 2016, 52, 341–351. [Google Scholar] [CrossRef]
- Shen, Z.; Ruan, Y.; Chao, X.; Zhang, J.; Li, R.; Shen, Q. Rhizosphere microbial community manipulated by 2 years of consecutive biofertilizer application associated with banana Fusarium wilt disease suppression. Biol. Fertil. Soils 2015, 51, 553–562. [Google Scholar] [CrossRef]
- Guo, Q.; Li, Y.; Lou, Y.; Shi, M.; Jiang, Y.; Zhou, J.; Sun, Y.; Xue, Q.; Lai, H. Bacillus amyloliquefaciens Ba13 induces plant systemic resistance and improves rhizosphere microecology against tomato yellow leaf curl virus disease. Appl. Soil Ecol. 2019, 137, 154–166. [Google Scholar] [CrossRef]
- Gowtham, H.; Murali, M.; Singh, S.B.; Lakshmeesha, T.; Murthy, K.N.; Amruthesh, K.; Niranjana, S. Plant growth promoting rhizobacteria-Bacillus amyloliquefaciens improves plant growth and induces resistance in chilli against anthracnose disease. Biol. Control 2018, 126, 209–217. [Google Scholar] [CrossRef]
- Cui, W.; He, P.; Munir, S.; He, P.; He, Y.; Li, X.; Yang, L.; Wang, B.; Wu, Y.; He, P. Biocontrol of soft rot of Chinese cabbage using an endophytic bacterial strain. Front. Microbiol. 2019, 10, 1471. [Google Scholar] [CrossRef] [Green Version]
- Sakarika, M.; Spanoghe, J.; Sui, Y.; Wambacq, E.; Grunert, O.; Haesaert, G.; Spiller, M.; Vlaeminck, S.E. Purple non-sulphur bacteria and plant production: Benefits for fertilization, stress resistance and the environment. Microb. Biotechnol. 2019. [Google Scholar] [CrossRef] [Green Version]
- Imam, S.; Noguera, D.R.; Donohue, T.J. Global insights into energetic and metabolic networks in Rhodobacter sphaeroides. BMC Syst. Biol. 2013, 7. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.L.; Zhang, G.M.; Li, X.K.; Wu, P.; Zhang, J. Enhancement of Rhodobacter sphaeroides growth and carotenoid production through biostimulation. J. Environ. Sci. 2015, 33, 21–28. [Google Scholar] [CrossRef]
- Kien, N.B.; Kong, I.S.; Lee, M.G.; Kim, J.K. Coenzyme Q(10) production in a 150-l reactor by a mutant strain of Rhodobacter sphaeroides. J. Ind. Microbiol. Biotechnol. 2010, 37, 521–529. [Google Scholar] [CrossRef]
- Kho, D.H.; Yoo, S.B.; Kim, J.S.; Kim, E.J.; Lee, J.K. Characterization of Cu- and Zn-containing superoxide dismutase of Rhodobacter sphaeroides. Fems Microbiol. Lett. 2004, 234, 261–267. [Google Scholar] [CrossRef]
- Holguin, G.; Vazquez, P.; Bashan, Y. The role of sediment microorganisms in the productivity, conservation, and rehabilitation of mangrove ecosystems: An overview. Biol. Fertil. Soils 2001, 33, 265–278. [Google Scholar] [CrossRef]
- Wu, P.; Zhang, Y.; Chen, Z.B.; Wang, Y.L.; Zhu, F.F.; Cao, B.; Wu, Y.; Li, N. The organophosphorus pesticides in soil was degradated by Rhodobacter sphaeroides after wastewater treatment. Biochem. Eng. J. 2019, 141, 247–251. [Google Scholar] [CrossRef]
- Koku, H.; Eroğlu, İ.; Gündüz, U.; Yücel, M.; Türker, L. Aspects of the metabolism of hydrogen production by Rhodobacter sphaeroides. Int. J. Hydrog. Energy 2002, 27, 1315–1329. [Google Scholar] [CrossRef]
- Kondo, K.; Nishihara, E.; Nakata, N. Effect of the Purple Non-Sulfur Bacterium (Rhodobacter sphaeroides) on the Fruit Quality of Tomato. In Proceedings of the XXVII International Horticultural Congress-IHC2006: International Symposium on Advances in Environmental Control, Automation 761, Seoul, Korea, 30 September 2007; pp. 581–587. [Google Scholar]
- Kensuke Kondo, N.N.a.E.N. Effect of Purple Non-sulfur Bacterium (Rhodobacter sphaeroides) Application on the Growth and Quality of Spinach and Komatsuna. Jpn. Soc. Agric. Technol. Manag. 2008, 14, 198–203. [Google Scholar]
- Hagemann, M.; Ribbeck-Busch, K.; Klaehn, S.; Hasse, D.; Steinbruch, R.; Berg, G. The plant-associated bacterium Stenotrophomonas rhizophila expresses a new enzyme for the synthesis of the compatible solute glucosylglycerol. J. Bacteriol. 2008, 190, 5898–5906. [Google Scholar] [CrossRef] [Green Version]
- Egamberdieva, D.; Kucharova, Z.; Davranov, K.; Berg, G.; Makarova, N.; Azarova, T.; Chebotar, V.; Tikhonovich, I.; Kamilova, F.; Validov, S.Z.; et al. Bacteria able to control foot and root rot and to promote growth of cucumber in salinated soils. Biol. Fertil. Soils 2011, 47, 197–205. [Google Scholar] [CrossRef]
- Alavi, P.; Starcher, M.R.; Zachow, C.; Muller, H.; Berg, G. Root-microbe systems: The effect and mode of interaction of Stress Protecting Agent (SPA) Stenotrophomonas rhizophila DSM14405(T). Front. Plant Sci. 2013, 4. [Google Scholar] [CrossRef] [Green Version]
- Wolf, A.; Fritze, A.; Hagemann, M.; Berg, G. Stenotrophomonas rhizophila sp nov., a novel plant-associated bacterium with antifungal properties. Int. J. Syst. Evol. Microbiol. 2002, 52, 1937–1944. [Google Scholar] [CrossRef]
- Schmidt, C.S.; Alavi, M.; Cardinale, M.; Müller, H.; Berg, G. Stenotrophomonas rhizophila DSM14405 T promotes plant growth probably by altering fungal communities in the rhizosphere. Biol. Fertil. Soils 2012, 48, 947–960. [Google Scholar] [CrossRef]
- Kai, M.; Effmert, U.; Berg, G.; Piechulla, B. Volatiles of bacterial antagonists inhibit mycelial growth of the plant pathogen Rhizoctonia solani. Arch. Microbiol. 2007, 187, 351–360. [Google Scholar] [CrossRef]
- Egamberdieva, D.; Jabborova, D.; Berg, G. Synergistic interactions between Bradyrhizobium japonicum and the endophyte Stenotrophomonas rhizophila and their effects on growth, and nodulation of soybean under salt stress. Plant Soil 2016, 405, 35–45. [Google Scholar] [CrossRef]
- Hu, L.; Robert, C.A.M.; Cadot, S.; Zhang, X.; Ye, M.; Li, B.; Manzo, D.; Chervet, N.; Steinger, T.; van der Heijden, M.G.A.; et al. Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 2018, 9, 2738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, J.; Moller, I. Percolation of Starch and Soluble Carbohydrates from Plant-Tissue for Quantitative-Determination with Anthrone. Anal. Biochem. 1975, 68, 87–94. [Google Scholar] [CrossRef]
- Van Dam, N.M.; Horn, M.; Mares, M.; Baldwin, I.T. Ontogeny constrains systemic protease inhibitor response in Nicotiana attenuata. J. Chem. Ecol. 2001, 27, 547–568. [Google Scholar] [CrossRef]
- Szeto, Y.T.; Tomlinson, B.; Benzie, I.F.F. Total antioxidant and ascorbic acid content of fresh fruits and vegetables: Implications for dietary planning and food preservation. Br. J. Nutr. 2002, 87, 55–59. [Google Scholar] [CrossRef] [Green Version]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; van Themaat, E.V.L.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef]
- Chakraborty, P.; Tribedi, P. Functional diversity performs a key role in the isolation of nitrogen-fixing and phosphate-solubilizing bacteria from soil. Folia Microbiol. 2019, 64, 461–470. [Google Scholar] [CrossRef]
- Glickmann, E.; Dessaux, Y. A Critical-Examination of the Specificity of the Salkowski Reagent for Indolic Compounds Produced by Phytopathogenic Bacteria. Appl. Environ. Microbiol. 1995, 61, 793–796. [Google Scholar] [CrossRef] [Green Version]
- Brian, C.; Louden, D.H.; Aaron, M.L. Use of Blue Agar CAS Assay for Siderophore Detection. J. Microbiol. Blol. Educ. 2011, 12, 51–53. [Google Scholar]
- O’Toole, G.A. Microtiter dish biofilm formation assay. JoVE (J. Vis. Exp.) 2011, 47, e2437. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335. [Google Scholar] [CrossRef] [Green Version]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Yong, X.; Zhang, R.; Zhang, N.; Chen, Y.; Huang, X.; Zhao, J.; Shen, Q. Development of a specific real-time PCR assay targeting the poly-gamma-glutamic acid synthesis gene, pgsB, for the quantification of Bacillus amyloliquefaciens in solid-state fermentation. Bioresour. Technol. 2013, 129, 477–484. [Google Scholar] [CrossRef]
- Guan, D.; Li, J.; Shen, D.; Cao, F.; Li, L.; Jiang, X. Identification and Quantification of Photosynthetic Bacteria by PCR Method. Chin. J. Appl. Environ. Biol. 2008, 14, 699–704. [Google Scholar] [CrossRef]
- Attard, E.; Poly, F.; Commeaux, C.; Laurent, F.; Terada, A.; Smets, B.F.; Recous, S.; Le Roux, X. Shifts between Nitrospira- and Nitrobacter-like nitrite oxidizers underlie the response of soil potential nitrite oxidation to changes in tillage practices. Environ. Microbiol. 2010, 12, 315–326. [Google Scholar] [CrossRef]
- Rosch, C.; Mergel, A.; Bothe, H. Biodiversity of denitrifying and dinitrogen-fixing bacteria in an acid forest soil. Appl. Environ. Microbiol. 2002, 68, 3818–3829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koper, T.E.; El-Sheikh, A.F.; Norton, J.M.; Klotz, M.G. Urease-encoding genes in ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 2004, 70, 2342–2348. [Google Scholar] [CrossRef] [Green Version]
- Alavi, P.; Starcher, M.R.; Thallinger, G.G.; Zachow, C.; Mueller, H.; Berg, G. Stenotrophomonas comparative genomics reveals genes and functions that differentiate beneficial and pathogenic bacteria. BMC Genom. 2014, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, Y.; Jiang, Y.H.; Yang, Y.F.; He, Z.L.; Luo, F.; Zhou, J.Z. Molecular ecological network analyses. BMC Bioinform. 2012, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Lu, G.; Yuan, M.; Yu, H.; Wang, S.; Li, X.; Deng, Y. Elevated temperature overrides the effects of N amendment in Tibetan grassland on soil microbiome. Soil Biol. Biochem. 2019, 136. [Google Scholar] [CrossRef]
- West, D.B. Introduction to Graph Theory; Prentice hall: Upper Saddle River, NJ, USA, 2001; Volume 2. [Google Scholar]
- Watts, D.J.; Strogatz, S.H. Collective dynamics of ‘small-world’ networks. Nature 1998, 393, 440. [Google Scholar] [CrossRef] [PubMed]
- Newman, M.E. Modularity and community structure in networks. Proc. Natl. Acad. Sci. USA 2006, 103, 8577–8582. [Google Scholar] [CrossRef] [Green Version]
- Krause, A.E.; Frank, K.A.; Mason, D.M.; Ulanowicz, R.E.; Taylor, W.W. Compartments revealed in food-web structure. Nature 2003, 426, 282–285. [Google Scholar] [CrossRef]
- Hayat, R.; Ali, S.; Amara, U.; Khalid, R.; Ahmed, I. Soil beneficial bacteria and their role in plant growth promotion: A review. Ann. Microbiol. 2010, 60, 579–598. [Google Scholar] [CrossRef]
- Di Salvo, L.P.; Cellucci, G.C.; Carlino, M.E.; de Salamone, I.E.G. Plant growth-promoting rhizobacteria inoculation and nitrogen fertilization increase maize (Zea mays L.) grain yield and modified rhizosphere microbial communities. Appl. Soil Ecol. 2018, 126, 113–120. [Google Scholar] [CrossRef]
- Goswami, D.; Thakker, J.N.; Dhandhukia, P.C. Portraying mechanics of plant growth promoting rhizobacteria (PGPR): A review. Cogent Food Agric. 2016, 2, 1127500. [Google Scholar] [CrossRef]
- Lundberg, D.S.; Teixeira, P.J. Root-exuded coumarin shapes the root microbiome. Proc. Natl. Acad. Sci. USA 2018, 115, 5629–5631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeAngelis, K.M.; Brodie, E.L.; DeSantis, T.Z.; Andersen, G.L.; Lindow, S.E.; Firestone, M.K. Selective progressive response of soil microbial community to wild oat roots. ISME J. 2009, 3, 168–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, S.; Richardson, A.E.; O’Callaghan, M.; DeAngelis, K.M.; Jones, E.E.; Stewart, A.; Firestone, M.K.; Condron, L.M. Effects of selected root exudate components on soil bacterial communities. FEMS Microbiol. Ecol. 2011, 77, 600–610. [Google Scholar] [CrossRef] [Green Version]
- Goldfarb, K.C.; Karaoz, U.; Hanson, C.A.; Santee, C.A.; Bradford, M.A.; Treseder, K.K.; Wallenstein, M.D.; Brodie, E.L. Differential growth responses of soil bacterial taxa to carbon substrates of varying chemical recalcitrance. Front. Microbiol. 2011, 2, 94. [Google Scholar] [CrossRef] [Green Version]
- Hou, J.; Liu, W.; Wang, B.; Wang, Q.; Luo, Y.; Franks, A.E. PGPR enhanced phytoremediation of petroleum contaminated soil and rhizosphere microbial community response. Chemosphere 2015, 138, 592–598. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; Van Themaat, E.V.L.; Schulze-Lefert, P. Structure and functions of the bacterial microbiota of plants. Ann. Rev. Plant. Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef] [Green Version]
- Bulgarelli, D.; Garrido-Oter, R.; Münch, P.C.; Weiman, A.; Dröge, J.; Pan, Y.; McHardy, A.C.; Schulze-Lefert, P. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 2015, 17, 392–403. [Google Scholar] [CrossRef] [Green Version]
- Müller, D.B.; Vogel, C.; Bai, Y.; Vorholt, J.A. The plant microbiota: Systems-level insights and perspectives. Annu. Rev. Genet. 2016, 50, 211–234. [Google Scholar] [CrossRef] [Green Version]
- Zhalnina, K.; Louie, K.B.; Hao, Z.; Mansoori, N.; da Rocha, U.N.; Shi, S.; Cho, H.; Karaoz, U.; Loqué, D.; Bowen, B.P. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 2018, 3, 470–480. [Google Scholar] [CrossRef] [Green Version]
- Hiltner, L. Über neuere erfahrungen und probleme auf dem debiete der bo denbakteriologie und unter besonderer berucksichtigung der grundund und brache. Zbl. Bakteriol. 1904, 2, 14–25. [Google Scholar]
- Hartmann, A.; Rothballer, M.; Schmid, M. Lorenz Hiltner, a pioneer in rhizosphere microbial ecology and soil bacteriology research. Plant Soil 2008, 312, 7–14. [Google Scholar] [CrossRef]
- Hiltner, L. Pflanzenschutz Nach Monaten Geordnet; Eine Anleitung für Landwirte, Gärtner, Obstbaumzüchter &c.; Eugen Ulmer: Stuttgart, Germany, 1909. [Google Scholar] [CrossRef] [Green Version]
- Edwards, J.A.; Santos-Medellín, C.M.; Liechty, Z.S.; Nguyen, B.; Lurie, E.; Eason, S.; Phillips, G.; Sundaresan, V. Compositional shifts in root-associated bacterial and archaeal microbiota track the plant life cycle in field-grown rice. PLoS Biol. 2018, 16, e2003862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, B.; Paulson, J.N.; Zheng, X.; Kolter, R. Simplified and representative bacterial community of maize roots. Proc. Natl. Acad. Sci. USA 2017, 114, E2450–E2459. [Google Scholar] [CrossRef] [Green Version]
- Layeghifard, M.; Hwang, D.M.; Guttman, D.S. Disentangling interactions in the microbiome: A network perspective. Trends Microbiol. 2017, 25, 217–228. [Google Scholar] [CrossRef]
- Buol, S.W.; Southard, R.J.; Graham, R.C.; McDaniel, P.A. Soil Genesis and Classification; John Wiley & Sons: Hoboken, NJ, USA, 2011. [Google Scholar]
- Vieira, S.; Sikorski, J.; Dietz, S.; Herz, K.; Schrumpf, M.; Bruelheide, H.; Scheel, D.; Friedrich, M.W.; Overmann, J. Drivers of the composition of active rhizosphere bacterial communities in temperate grasslands. ISME J. 2020, 14, 463–475. [Google Scholar] [CrossRef] [Green Version]
- Prasad, A.A.; Babu, S. Compatibility of Azospirillum brasilense and Pseudomonas fluorescens in growth promotion of groundnut (Arachis hypogea L.). An. Acad. Bras. Ciênc. 2017, 89, 1027–1040. [Google Scholar] [CrossRef] [Green Version]
- Borges, A.; Abreu, A.C.; Dias, C.; Saavedra, M.J.; Borges, F.; Simões, M. New perspectives on the use of phytochemicals as an emergent strategy to control bacterial infections including biofilms. Molecules 2016, 21, 877. [Google Scholar] [CrossRef]
- Khan, Z.; Roman, D.; Kintz, T.; delas Alas, M.; Yap, R.; Doty, S. Degradation, phytoprotection and phytoremediation of phenanthrene by endophyte Pseudomonas putida, PD1. Environ. Sci. Technol. 2014, 48, 12221–12228. [Google Scholar] [CrossRef]
- Rodríguez, H.; Fraga, R. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 1999, 17, 319–339. [Google Scholar] [CrossRef]
- Arora, N.K.; Verma, M.; Mishra, J. Rhizobial bioformulations: Past, present and future. In Rhizotrophs: Plant Growth Promotion to Bioremediation; Springer: Cham, Switzerland, 2017; pp. 69–99. [Google Scholar]
- Soltani, A.-A.; Khavazi, K.; Asadi-Rahmani, H.; Omidvari, M.; Dahaji, P.A.; Mirhoseyni, H. Plant growth promoting characteristics in some Flavobacterium spp. isolated from soils of Iran. J. Agric. Sci. 2010, 2, 106. [Google Scholar] [CrossRef] [Green Version]
- Mawdsley, J.L.; Burns, R.G. Inoculation of plants with a Flavobacterium species results in altered rhizosphere enzyme activities. Soil Biol. Biochem. 1994, 26, 871–882. [Google Scholar] [CrossRef]
- Peiffer, J.A.; Spor, A.; Koren, O.; Jin, Z.; Tringe, S.G.; Dangl, J.L.; Buckler, E.S.; Ley, R.E. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. USA 2013, 110, 6548–6553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, S.; Nuccio, E.; Herman, D.J.; Rijkers, R.; Estera, K.; Li, J.; da Rocha, U.N.; He, Z.; Pett-Ridge, J.; Brodie, E.L. Successional trajectories of rhizosphere bacterial communities over consecutive seasons. mBio 2015, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, S.; Fen, L.; Guo, C.; Wang, T.L.; Zhang, J.; Deng, L.; Luo, K.; Xiang, L.Y.; Huang, H.; Mei, X. Negative plant-soil feedback driven by re-assemblage of the rhizosphere microbiome with the growth of Panax notoginseng. Front. Microbiol. 2019, 10, 1597. [Google Scholar]
- Mendes, L.W.; Kuramae, E.E.; Navarrete, A.A.; Van Veen, J.A.; Tsai, S.M. Taxonomical and functional microbial community selection in soybean rhizosphere. ISME J. 2014, 8, 1577–1587. [Google Scholar] [CrossRef]
- Fierer, N.; Lauber, C.L.; Ramirez, K.S.; Zaneveld, J.; Bradford, M.A.; Knight, R. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 2012, 6, 1007–1017. [Google Scholar] [CrossRef] [Green Version]
- Madigan, M.T. Microbiology of nitrogen fixation by anoxygenic photosynthetic bacteria. In Anoxygenic Photosynthetic Bacteria; Springer: Cham, Switzerland, 1995; pp. 915–928. [Google Scholar]
- Sorokin, D.Y.; Lücker, S.; Vejmelkova, D.; Kostrikina, N.A.; Kleerebezem, R.; Rijpstra, W.I.C.; Damsté, J.S.S.; Le Paslier, D.; Muyzer, G.; Wagner, M. Nitrification expanded: Discovery, physiology and genomics of a nitrite-oxidizing bacterium from the phylum Chloroflexi. ISME J. 2012, 6, 2245–2256. [Google Scholar] [CrossRef] [Green Version]
- Prasanna, R.; Bidyarani, N.; Babu, S.; Hossain, F.; Shivay, Y.S.; Nain, L. Cyanobacterial inoculation elicits plant defense response and enhanced Zn mobilization in maize hybrids. Cogent Food Agric. 2015, 1, 998507. [Google Scholar] [CrossRef]
- Karthikeyan, N.; Prasanna, R.; Nain, L.; Kaushik, B.D. Evaluating the potential of plant growth promoting cyanobacteria as inoculants for wheat. Eur. J. Soil Biol. 2007, 43, 23–30. [Google Scholar] [CrossRef]
- Rai, A.N.; Bergman, B.; Rasmussen, U. Cyanobacteria in Symbiosis; Springer: Cham, Switzerland, 2002. [Google Scholar]
- Sharma, N.K.; Rai, A.K.; Stal, L.J. Cyanobacteria: An. Economic Perspective; John Wiley & Sons: Hoboken, NJ, USA, 2013. [Google Scholar]
- Prasanna, R.; Sood, A.; Rath, S.; Singh, P.K. Cyanobacteria as a green option for sustainable agriculture. In Cyanobacteria: An Economic Perspective; Wiley Online Library: London, UK, 2014; pp. 145–166. [Google Scholar]
- Rossi, F.; Li, H.; Liu, Y.; De Philippis, R. Cyanobacterial inoculation (cyanobacterisation): Perspectives for the development of a standardized multifunctional technology for soil fertilization and desertification reversal. Earth Sci. Rev. 2017, 171, 28–43. [Google Scholar] [CrossRef]
- Klatt, C.G.; Liu, Z.F.; Ludwig, M.; Kuhl, M.; Jensen, S.I.; Bryant, D.A.; Ward, D.M. Temporal metatranscriptomic patterning in phototrophic Chloroflexi inhabiting a microbial mat in a geothermal spring. ISME J. 2013, 7, 1775–1789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bateson, M.M.; Ward, D.M. Photoexcretion and fate of glycolate in a hot spring cyanobacterial mat. Appl. Environ. Microbiol. 1988, 54, 1738–1743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, K.L.; Tayne, T.A.; Ward, D.M. Formation and fate of fermentation products in hot spring cyanobacterial mats. Appl. Environ. Microbiol. 1987, 53, 2343–2352. [Google Scholar] [CrossRef] [Green Version]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef]
- Lu, L.; Xing, D.; Ren, Z.J. Microbial community structure accompanied with electricity production in a constructed wetland plant microbial fuel cell. Bioresour. Technol. 2015, 195, 115–121. [Google Scholar] [CrossRef]
- Sellstedt, A.; Richau, K.H. Aspects of nitrogen-fixing Actinobacteria, in particular free-living and symbiotic Frankia. FEMS Microbiol. Lett. 2013, 342, 179–186. [Google Scholar] [CrossRef] [Green Version]
- Strap, J.L. Actinobacteria–plant interactions: A boon to agriculture. In Bacteria in Agrobiology: Plant Growth Responses; Springer: Cham, Switzerland, 2011; pp. 285–307. [Google Scholar]
- Hamedi, J.; Mohammadipanah, F. Biotechnological application and taxonomical distribution of plant growth promoting actinobacteria. J. Ind. Microbiol. Biotechnol. 2015, 42, 157–171. [Google Scholar] [CrossRef]
- Ahn, J.-H.; Song, J.; Kim, B.-Y.; Kim, M.-S.; Joa, J.-H.; Weon, H.-Y. Characterization of the bacterial and archaeal communities in rice field soils subjected to long-term fertilization practices. J. Microbiol. 2012, 50, 754–765. [Google Scholar] [CrossRef]
- Diamond, S.; Andeer, P.F.; Li, Z.; Crits-Christoph, A.; Burstein, D.; Anantharaman, K.; Lane, K.R.; Thomas, B.C.; Pan, C.; Northen, T.R. Mediterranean grassland soil C–N compound turnover is dependent on rainfall and depth, and is mediated by genomically divergent microorganisms. Nat. Microbiol. 2019, 4, 1356–1367. [Google Scholar] [CrossRef] [Green Version]
- Finkel, O.M.; Castrillo, G.; Paredes, S.H.; González, I.S.; Dangl, J.L. Understanding and exploiting plant beneficial microbes. Curr. Opin. Plant. Biol. 2017, 38, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Pii, Y.; Mimmo, T.; Tomasi, N.; Terzano, R.; Cesco, S.; Crecchio, C. Microbial interactions in the rhizosphere: Beneficial influences of plant growth-promoting rhizobacteria on nutrient acquisition process. A review. Biol. Fertil. Soils 2015, 51, 403–415. [Google Scholar] [CrossRef]
- Kuypers, M.M.M.; Marchant, H.K.; Kartal, B. The microbial nitrogen-cycling network. Nat. Rev. Microbiol 2018, 16, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Xu, J.; Kloepper, J.W. Plant-microbe-soil fertility interaction impacts performance of a Bacillus-containing bioproduct on bell pepper. J. Basic Microbiol. 2020, 60, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Fisher, K.A.; Yarwood, S.A.; James, B.R. Soil urease activity and bacterial ureC gene copy numbers: Effect of pH. Geoderma 2017, 285, 1–8. [Google Scholar] [CrossRef]
- Leghari, S.J.; Wahocho, N.A.; Laghari, G.M.; HafeezLaghari, A.; MustafaBhabhan, G.; HussainTalpur, K.; Bhutto, T.A.; Wahocho, S.A.; Lashari, A.A. Role of nitrogen for plant growth and development: A review. Adv. Environ. Biol. 2016, 10, 209–219. [Google Scholar]
- Hesheng, L. Principles and Techniques of Plant. In Physiological and Biochemical Experiments; Higher Education Press: Beijing, China, 2000. [Google Scholar]





| Species | Primer Sequence | Origin | PCR Product | Reference |
|---|---|---|---|---|
| S. rhizophila | TCTCAACCTGGGTACCGTAATA | rpfX | 87-bp | This study |
| AGATGTCCAGGCAACAGTTC | ||||
| R. sphaeroides | GCCTCGGCCAAGACCAACC | gyr B | 250-bp | [51] |
| GCTCGCCGGTGATGAAGATGGG | ||||
| B. amyloliquefaciens | TGGCGCCATGAGAATCCT | pgs B | 66-bp | [50] |
| GCAAAGCCGTTTACGAAATGA | ||||
| _ | CAGACCGACGTGTGCGAAAG | nxrA | 320-bp | [52] |
| TCCACAAGGAACGGAAGGTC | ||||
| _ | AAAGGYGGWATCGGYAARTCCACCAC | nifH | 457-bp | [53] |
| TTGTTSGCSGCRTACATSGCCATCAT | ||||
| _ | TGGGCCTTAAAATHCAYGARGAYTGGG | uerC | 327-bp | [54] |
| GGTGGTGGCACACCATNANCATRTC |
| Plants | Statistics § | ||||||
|---|---|---|---|---|---|---|---|
| Sr | Rs | Ba | CK | Sr vs. CK | Rs vs. CK | Ba vs. CK | |
| Chlorophyll # | 42.78 ± 1.14 | 39.59 ± 0.87 | 45.66 ± 1.27 | 42.29 ± 0.96 | ns | ns | ns |
| Biomass (g) † | 1.17 ± 0.07 | 1.35 ± 0.11 | 1.35 ± 0.07 | 1.08 ± 0.09 | ns | ns | ns |
| Soluble starch (mg.g−1) † | 11.22 ± 0.32 | 9.89 ± 0.28 | 12.32 ± 0.15 | 10.18 ± 0.95 | ns | ns | ns |
| Soluble protein (mg.g−1) † | 24.25 ± 0.30 | 22.59 ± 0.54 | 26.47 ± 0.17 | 21.92 ± 0.42 |  * * | ns |  ** ** |
| Vitamin C (mg.g−1) † | 0.43 ± 0.02 | 0.42 ± 0.03 | 0.43 ± 0.02 | 0.47 ± 0.02 | ns | ns | ns |
| TC (g.kg−1) † | 301.51 ± 0.80 | 271.67 ± 3.59 | 300.65 ± 0.65 | 306.78 ± 1.55 | ns | ***  | ns |
| TN (g.kg−1) † | 53.30 ± 1.59 | 64.11 ± 0.56 | 66.93 ± 3.02 | 47.86 ± 0.13 |  ** ** |  *** *** |  *** *** |
| TS (g.kg−1) † | 9.68 ± 0.10 | 10.40 ± 0.10 | 10.95 ± 0.25 | 10.79 ± 0.35 | ns | ns | ns |
 represents increase,
represents increase,  represents decrease.
represents decrease.| Sr | Rs | Ba | CK | Statistics § | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rhizosphere | Bulk Soil | Rhizosphere | Bulk Soil | Rhizosphere | Bulk Soil | Rhizosphere | Bulk Soil | Sr Rhizosphere vs. Bulk | Rs Rhizosphere vs. Bulk | Ba Rhizosphere vs. Bulk | CK Rhizosphere vs. Bulk | Sr vs. CK rhizosphere | Rs vs. CK rhizosphere | Ba vs. CK rhizosphere | |
| pH ∫ | 7.71 ± 0.13 | 7.50 ± 0.09 | 7.67 ± 0.13 | 7.52 ± 0.05 | 7.55 ± 0.10 | 7.30 ± 0.15 | 7.49 ± 0.03 | 7.45 ± 0.03 | ns | ns | ns | ns | ns | ns | ns |
| Ca (mg.g−1) ∫ | 2923.44 ± 17.42 | 2775.27 ± 34.63 | 3020.09 ± 55.99 | 3031.11 ± 41.29 | 2697.36 ± 30.48 | 3138.69 ± 48.92 | 3159.29 ± 37.36 | 3218.87 ± 20.09 | ns | ns | ***  | ns | *  | ns | ***  |
| Mg (mg.g−1) ∫ | 684.41 ± 20.36 | 589.71 ± 23.29 | 828.84 ± 38.11 | 909.06 ± 12.27 | 559.76 ± 23.44 | 850.69 ± 28.21 | 816.57 ± 29.05 | 855.39 ± 10.84 | ns | ns | ***  | ns | **  | ns | ***  |
| Fe (mg.g−1) ∫ | 2251.47 ± 16.43 | 2312.01 ± 17.51 | 2419.31 ± 58.83 | 2422.01 ± 33.97 | 2124.92 ± 38.91 | 2473.94 ± 33.77 | 2365.15 ± 24.4 | 2473.95 ± 8.06 | ns | ns | ***  | ns | ns | ns | ***  |
| Mn (mg.g−1) ∫ | 59.44 ± 0.31 | 60.16 ± 0.28 | 64.72 ± 2.75 | 62.36 ± 0.67 | 55.32 ± 1.02 | 62.92 ± 0.37 | 63.05 ± 0.96 | 65.15 ± 1.22 | ns | ns | **  | ns | ns | ns | **  |
| Al (mg.g−1) ∫ | 1021.41 ± 10.44 | 968.59 ± 5.58 | 1032.47 ± 9.91 | 1028.72 ± 7.6 | 999.21 ± 7.97 | 1048.67 ± 6.38 | 1018.19 ± 7.39 | 1035.32 ± 3.6 |  *** *** | ns | **  | ns | ns | ns | ns |
| K (mg.g−1) ∫ | 1784.97 ± 17.7 | 1622.75 ± 15.68 | 1917.26 ± 41.16 | 1790.56 ± 21.41 | 1728.42 ± 49.49 | 1838.31 ± 19.79 | 1839.89 ± 18.98 | 1878.54 ± 13.7 |  ** ** |  * * | ns | ns | ns | ns | ns |
| P (mg.g−1) ∫ | 250.85 ± 3.93 | 201.7 ± 6.12 | 280.34 ± 17.47 | 242.94 ± 7.69 | 211.48 ± 10.01 | 188.09 ± 9.17 | 202.21 ± 6.57 | 168.02 ± 1.7 |  ** ** | ns | ns | ns |  * * |  *** *** | ns |
| NH4+ (mg.kg−1) ∫ | 20.94 ± 2.63 | 3.65 ± 0.08 | 12.95 ± 1.52 | 3.69 ± 0.36 | 11.21 ± 1.22 | 4.62 ± 0.12 | 25.79 ± 1.08 | 4.47 ± 0.1 |  *** *** |  *** *** |  * * |  *** *** | ns | *  | **  |
| NO3− (mg.kg−1) ∫ | 146.92 ± 3.37 | 90.26 ± 2.48 | 115.48 ± 7.99 | 107.74 ± 16.58 | 90.43 ± 6.00 | 146.34 ± 13.17 | 156.09 ± 4.99 | 149.84 ± 3.37 |  ** ** | ns | **  | ns | ns | **  | *  |
| TC (g.kg−1) ∫ | 21.51 ± 0.82 | 20.42 ± 0.3 | 28.53 ± 2.9 | 22.26 ± 0.33 | 20.99 ± 0.53 | 20.8 ± 0.27 | 23.85 ± 0.46 | 19.73 ± 0.54 | ns |  ** ** | ns | ns | ns | ns | ns |
| TN (g.kg−1) ∫ | 1.45 ± 0.09 | 1.35 ± 0.04 | 2 ± 0.22 | 1.6 ± 0.07 | 1.43 ± 0.07 | 1.48 ± 0.06 | 1.51 ± 0.14 | 0.98 ± 0.15 | ns | ns | ns | ns | ns | ns | ns |
| TS (g.kg−1) ∫ | 0.33 ± 0.02 | 0.52 ± 0.02 | 0.41 ± 0.05 | 0.57 ± 0.02 | 0.32 ± 0.02 | 0.44 ± 0.03 | 0.33 ± 0.02 | 0.41 ± 0.03 | ***  | **  | *  | ns | ns | ns | ns |
 represents increase,
represents increase,  represents decrease.
represents decrease.Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Gao, J.; Bai, Z.; Wu, S.; Li, X.; Wang, N.; Du, X.; Fan, H.; Zhuang, G.; Bohu, T.; et al. Unraveling Mechanisms and Impact of Microbial Recruitment on Oilseed Rape (Brassica napus L.) and the Rhizosphere Mediated by Plant Growth-Promoting Rhizobacteria. Microorganisms 2021, 9, 161. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms9010161
Liu Y, Gao J, Bai Z, Wu S, Li X, Wang N, Du X, Fan H, Zhuang G, Bohu T, et al. Unraveling Mechanisms and Impact of Microbial Recruitment on Oilseed Rape (Brassica napus L.) and the Rhizosphere Mediated by Plant Growth-Promoting Rhizobacteria. Microorganisms. 2021; 9(1):161. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms9010161
Chicago/Turabian StyleLiu, Ying, Jie Gao, Zhihui Bai, Shanghua Wu, Xianglong Li, Na Wang, Xiongfeng Du, Haonan Fan, Guoqiang Zhuang, Tsing Bohu, and et al. 2021. "Unraveling Mechanisms and Impact of Microbial Recruitment on Oilseed Rape (Brassica napus L.) and the Rhizosphere Mediated by Plant Growth-Promoting Rhizobacteria" Microorganisms 9, no. 1: 161. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms9010161