The Use of Probiotics to Fight Biofilms in Medical Devices: A Systematic Review and Meta-Analysis
Abstract
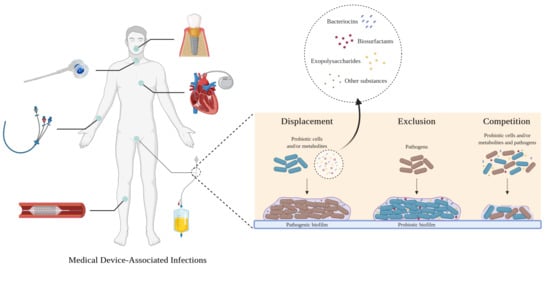
:1. Introduction
2. Methods
2.1. Search Strategy, Study Eligibility and Data Extraction
2.2. Quality Assessment
2.3. Meta-Analysis
3. Results and Discussion
3.1. Study Selection and Characterization
| Antibiofilm Substances and Probiotic Strains | Abiotic Surface | Biofilm Forming Pathogens | Percentages of Reduction | Ref. | Major Conclusions | |
|---|---|---|---|---|---|---|
| Biosurfactants | L. brevis L. gasseri L. jensenii L. rhamnosus | Polystyrene Silicone elastomeric discs | Ac. baumannii C. albicans C. krusei C. tropicalis En. aerogenes E. coli K. pneumoniae S. aureus S. saprophyticus | 58% 37% 37% 33% 64% 46%–65% 16% 61% 39% | [27] [57,80] [80] [80] [80] [27,80] [80] [27] [80] | Biosurfactants disrupted the biofilms of all bacteria by 16%–65%, depending on the concentration. For yeasts, a biofilm reduction of 35% was achieved. |
| Bacteriocins | L. acidophilus L. plantarum | Foley silicone catheter pieces Polystyrene | P. aeruginosa Ser. marcescens | 59% 48% | [84] [85] | Bacteriocins showed inhibitory activity against P. aeruginosa (59%) and living cells of Ser. marcescens (48%). |
| EPS | Leu. citreum Leu. mesenteroides Leu. pseudo-mesenteroides Ped. pentosaceus | N.A. | Ent. faecalis E. coli S. aureus | 53% 62% 77% | [86] [86] [86] | The capacity of EPS to disrupt pre-formed biofilms increased with increasing concentrations, and it was lower than the capacity to prevent adhesion. Biofilm formation was reduced by 53%–77%. |
| Cell-free supernatants | L. fermentum L. gasseri L. helveticus L. pentosus L. plantarum L. rhamnosus Strep. salivarius | Glass Polystyrene Polyurethane PVC | C. albicans C. krusei C. parapsilosis C. tropicalis E. coli K. pneumoniae P. aeruginosa S. aureus | 80% 67% 40% 64% N.A. 78% 74% 50% | [87] [88] [88] [88] [89] [90] [90] [91] | CFS induced biofilm disruption on the different surfaces by 38%–80%, depending on the species. The neutralized supernatants inhibited P. aeruginosa (74%) and K. pneumoniae biofilm formation (78%). |
| Cells | B. infantis B. longum Ent. faecium L. acidophilus L. casei L. casei rhamnosus L. casei shirota L. fermentum L. helveticus L. paracasei L. plantarum L. reuteri L. rhamnosus L. rhamnosus GG Lact. lactis Lact. lactis cremoris Strep. cremoris Strep. salivarius Strep. thermophilus | Bovine enamel saliva-coated Denture surface Glass Polyurethane Saliva-conditioned titanium discs Silicone latex Silicone rubber | At. vaginae C. albicans C. tropicalis E. coli G. vaginalis S. aureus Strep. mutans Strep. oralis Staphylococcal strains Streptococcal strains | N.A. 80%–99% 88%–95% 80% N.A. 98% 29%–99% 99% 83% 83% | [89] [60,65,87,92] [60,92] [93] [89] [94] [95,96] [96] [60] [60] | Probiotics overlaid on pre-formed biofilms reduced the biofilm culturable cells of Gram-positive bacteria by 79%–99% and biofilm formation by 89%–94%. Biofilm culturable cells of yeasts were reduced by more than 63%. B. infantis and Ent. faecium did not reduce the number of yeasts in biofilms. L. rhamnosus microcapsules reduced E. coli culturable cells in the biofilm up to 80%, in a dose-dependent manner. |
| Lipoteichoic acid (LTA) | L. plantarum | Glass Polystyrene | A. naeslundii Ent. faecalis L. salivarius Strep. mutans | N.A. N.A. N.A. N.A. | [97] [97] [97] [97,98] | LTA activity was inconsistent. |
| Antibiofilm Substances and Probiotic Strains | Abiotic Surface | Biofilm Forming Pathogens | Percentages of Reduction | Ref. | Major Conclusions | |
|---|---|---|---|---|---|---|
| Biosurfactants | L. acidophilus L. brevis L. casei L. delbrueckii L. fermentum L. helveticus L. paracasei L. plantarum L. reuteri L. rhamnosus Lact. lactis Strep. thermophilus | PDMS discs Polystyrene Silicone elastomeric discs Silicone rubber | Bac. cereus Bac. subtilis C. albicans C. tropicalis Ent. faecalis E. coli K. pneumoniae Lis. innocua Lis. monocytogenes Pr. mirabilis Pr. vulgaris Prov. stuartii P. aeruginosa P. putida R. dentocariosa Sal. typhi Ser. marcescens Sh. flexneri S. aureus S. epidermidis Strep. salivarius | 87% 79% 50%–85% 56%–67% N.A. 50%–59% N.A. 82% 84% N.A. 65%–75% N.A. 49%–70% 65% 78%–89% 56% 60% 40% 61%–96% 85%–94% 90%–93% | [81] [99] [57,62,100,101] [61,62] [101] [81,99,101] [101] [81] [81] [101] [99,100] [101] [81,99,101] [99] [61,62] [81] [102] [81] [61,62,81,99,100] [61,62,81,101] [61,62] | Pre-adsorbed biosurfactants displayed high anti-adhesive activity against both Gram-positive (61–97%) and Gram-negative (40%–75%) bacteria. Pre-adsorbed biosurfactant reduced the adhesion of yeasts to silicone by 50%–85%. |
| Bacteriocins | L. fermentum L. plantarum | Foley silicone catheter pieces Polystyrene | P. aeruginosa S. aureus | 99% N.A. | [103] [59,103] | Pre-coating with bacteriocins reduced the number of biofilm culturable cells by 99%. |
| EPS | L. fermentum Leu. citreum Leu. mesenteroides Leu. pseudo-mesenteroides Ped. pentosaceus | Polystyrene | Ent. faecalis E. coli P. aeruginosa S. aureus | 88% 90% 96% 87% | [86] [86] [103] [86] | Pre-coating with EPS reduced the number of biofilm culturable cells of P. aeruginosa by 96% and inhibited the adhesion of bacteria in a dose-dependent manner (87%–90%). |
| Cells | E. coli Nissle 1917 L. acidophilus L. casei L. casei rhamnosus L. fermentum L. paracasei L. rhamnosus Lact. lactis Lact. lactis ssp. lactis Strep. thermophilus | Denture surface Foley silicone catheter pieces Glass Saliva-coated hydroxyapatite discs Polystyrene Saliva-conditioned titanium discs Silicone Silicone latex | A. naeslundii C. albicans Ent. faecalis E. coli F. nucleatum Klebsiella ssp. Non-mutans streptococci strains P. aeruginosa S. aureus Strep. mutans Strep. oralis Strep. sobrinus V. dispar | 33% 99% 99% 99% 60% N.A. 8% N.A. 99% 30%-99% 79%-99% 89% 68% | [104] [65] [12] [105] [104] [105] [106] [105] [94,105,107] [96,106] [96,104] [104] [104] | Probiotics reduced the adhesion of pathogens up to 3 Log CFU and biofilm biomass by 8%–30%. Pre-coating with EcN biofilms reduced the adherence of Ent. faecalis on silicone up to 2 Log CFU. |
| Collagen-binding protein (p29) | L. fermentum | Polyisobutylene-polystyrene (PIB-PS) copolymerSilicone rubber | Ent. faecalis E. coli | 47% 75% | [58] [58] | Coating with p29 resulted in a reduction of 34% and 75% in E. coli adhesion, and 47% and 18% in Ent. faecalis adhesion to silicone rubber and PIB-PS, respectively. |
| Lipoteichoic acid (LTA) | L. plantarum | Polystyrene | Strep. mutans | 40% | [98] | Biofilm formation was inhibited, but to a lesser degree in comparison with co-incubation. |
| Antibiofilm Substances and Probiotic Strains | Abiotic Surface | Biofilm Forming Pathogens | Percentages of Reduction | Ref. | Major Conclusions | |
|---|---|---|---|---|---|---|
| Biosurfactants | L. acidophilus L. brevis L. helveticus L. jensenii L. paracasei L. reuteri L. rhamnosus | Medical grade silicone tubes Polystyrene Polystyrene pre-coated with human plasma Silicone elastomeric discs Saliva-conditioned titanium discs | Ac. Baumannii Bac. cereus C. albicans E. coli P. aeruginosa Ser. marcescens S. aureus Strep. mutans Strep. oralis | 76% 100% 90%–100% 79%–100% 100% 73% 88%–100% 99% 99% | [27] [81] [57,81] [27,81] [81] [102] [27,81] [64] [64] | Biosurfactants displayed high anti-adhesive activity and reduced biofilm biomass by 60%–100% and culturable cells by 90%–99%. The inhibitory effect was dose-dependent. |
| Bacteriocins | L. fermentum L. plantarum | Polystyrene | P. aeruginosa S. aureus | 56%–93% 62% | [59,103] [59] | Co-incubation with bacteriocins reduced the number of P. aeruginosa culturable cells by 93% and biofilm formation of pathogens by 56%–62%. |
| EPS | L. delbrueckii ssp. bulgaricus L. fermentum L. rhamnosus | Polystyrene | Bac. cereus Ent. faecalis Lis. monocytogenes P. aeruginosa | 90% 87% 88% 88%–97% | [108] [108] [108] [103,108] | Co-incubation with EPS reduced the number of P. aeruginosa culturable cells by 97% and inhibited biofilm formation between 74% and 90%, depending on the species, in a dose-dependent manner. |
| Cell-free supernatants | Bac. subtilis L. acidophilus L. fermentum L. gasseri L. helveticus L. paracasei L. plantarum L. rhamnosus Strep. salivarius | Glass Polystyrene Polyurethane PVC Saliva-conditioned titanium discs Silicone | C. albicans C. krusei C. parapsilosis C. tropicalis Ent. faecalis E. coli K. pneumoniae P. aeruginosa S. aureus Strep. mutans Strep. oralis | 90% 71% 41% 67% 61% 63% 99% 57% 57%–99% 53%–99% 99% | [87] [88] [88] [88] [109] [109] [110] [91] [91,111] [96,98,112] [96] | CFS were able to reduce the number of biofilm cells by more than 81% and inhibit the ability of pathogens to adhere to the different surfaces by 39–99%. Neutralized supernatants had less effect on biofilm formation. |
| Cells | E. coli Nissle 1917 L. acidophilus L. casei L. casei rhamnosus L. fermentum L. helveticus L. paracasei L. plantarum L. rhamnosus L. rhamnosus GG L. salivarius Lact. lactis ssp. lactis Strep. thermophilus | Bovine enamel saliva-coated Glass Polystyrene Polyurethane Polypropylene Saliva-coated hydroxyapatite discs Saliva-conditioned titanium discs Silicone latex Silicone rubber | A. naeslundii C. albicans E. coli F. nucleatum K. pneumoniae Non-mutans streptococci strains P. aeruginosa S. aureus S. epidermidis Strep. mutans Strep. oralis Strep. sanguinisStrep. sobrinus V. díspar | 22% 53%–72% 82%–93% 55% 99% 11% N.A. 99% 99% 9%–99% 65%–99% N.A. 76% 32% | [104] [63,87] [93,113,114] [104] [110] [106] [113] [94,113] [113] [63,95,96,106,112] [96,104] [63] [104] [104] | The adhesion of pathogens was reduced by the presence of probiotic cells (11%–93%), and their culturability decreased up to 7.2 Log CFU. L. rhamnosus microcapsules reduced biofilm formation up to 82% in a dose-dependent manner. Lactobacillus strains inhibited the growth of an uropathogenic biofilm on silicone rubber for at least 8 days. EcN was able to outcompete pathogenic strains during biofilm formation, reducing culturability up to 4 Log. |
| Lipoteichoic acid (LTA) | L. plantarum | Glass Human dentin slices Polystyrene Saliva-coated hydroxyapatite discs | A. naeslundii Ent. faecalis L. salivarius Strep. mutans | 57% 57% 57% 57%–75% | [97] [97] [97] [97,98] | LTA inhibited single- and multi-species biofilm formation by 75% and 57%, respectively. |
3.2. Quality Assessment
3.3. Meta-Analysis
3.3.1. Publication Bias
3.3.2. Limitations and Strengths
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Vertes, A.; Hitchins, V.; Phillips, K.S. Analytical Challenges of Microbial Biofilms on Medical Devices. Anal. Chem. 2012, 84, 3858–3866. [Google Scholar] [CrossRef] [PubMed]
- Khatoon, Z.; McTiernan, C.D.; Suuronen, E.J.; Mah, T.-F.; Alarcon, E.I. Bacterial biofilm formation on implantable devices and approaches to its treatment and prevention. Heliyon 2018, 4, e01067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azevedo, A.S.; Almeida, C.; Melo, L.F.; Azevedo, N.F. Impact of polymicrobial biofilms in catheter-associated urinary tract infections. Crit. Rev. Microbiol. 2016, 43, 423–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singha, P.; Locklin, J.; Handa, H. A review of the recent advances in antimicrobial coatings for urinary catheters. Acta Biomater. 2017, 50, 20–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reid, G. Biofilms in infectious disease and on medical devices. Int. J. Antimicrob. Agents 1999, 11, 223–226. [Google Scholar] [CrossRef]
- Chen, M.; Yu, Q.; Sun, H. Novel Strategies for the Prevention and Treatment of Biofilm Related Infections. Int. J. Mol. Sci. 2013, 14, 18488–18501. [Google Scholar] [CrossRef] [Green Version]
- Talsma, S.S. Biofilms on Medical Devices. Home Health Nurse J. Home Care Hosp. Prof. 2007, 25, 589–594. [Google Scholar] [CrossRef]
- Darouiche, R.O. Device-Associated Infections: A Macroproblem that Starts with Microadherence. Clin. Infect. Dis. 2001, 33, 1567–1572. [Google Scholar] [CrossRef]
- Tenke, P.; Köves, B.; Nagy, K.; Hultgren, S.J.; Mendling, W.; Wullt, B.; Grabe, M.; Wagenlehner, F.M.E.; Cek, M.; Pickard, R.; et al. Update on biofilm infections in the urinary tract. World J. Urol. 2012, 30, 51–57. [Google Scholar] [CrossRef] [Green Version]
- Percival, S.L.; Suleman, L.; Vuotto, C.; Donelli, G. Healthcare-associated infections, medical devices and biofilms: Risk, tolerance and control. J. Med. Microbiol. 2015, 64, 323–334. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Z.; Wang, Z.; Li, S.; Yuan, X. Antimicrobial strategies for urinary catheters. J. Biomed. Mater. Res. Part A 2019, 107, 445–467. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zhu, Z.; Wang, J.; Lopez, A.I.; Li, S.; Kumar, A.; Yu, F.; Chen, H.; Cai, C.; Zhang, L. Probiotic E. coli Nissle 1917 biofilms on silicone substrates for bacterial interference against pathogen colonization. Acta Biomater. 2017, 50, 353–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramstedt, M.; Ribeiro, I.A.C.; Bujdakova, H.; Mergulhão, F.J.M.; Jordao, L.; Thomsen, P.; Alm, M.; Burmølle, M.; Vladkova, T.; Can, F.; et al. Evaluating Efficacy of Antimicrobial and Antifouling Materials for Urinary Tract Medical Devices: Challenges and Recommendations. Macromol. Biosci. 2019, 19, e1800384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donlan, R. Biofilms and Device-Associated Infections. Emerg. Infect. Dis. 2001, 7, 277–281. [Google Scholar] [CrossRef]
- Siddiq, D.M.; Darouiche, R.O. New strategies to prevent catheter-associated urinary tract infections. Nat. Rev. Urol. 2012, 9, 305–314. [Google Scholar] [CrossRef]
- Veerachamy, S.; Yarlagadda, T.; Manivasagam, G.; Yarlagadda, P.K. Bacterial adherence and biofilm formation on medical implants: A review. Proc. Inst. Mech. Eng. Part H J. Eng. Med. 2014, 228, 1083–1099. [Google Scholar] [CrossRef]
- Rabin, N.; Zheng, Y.; Opoku-Temeng, C.; Du, Y.; Bonsu, E.; Sintim, H.O. Biofilm formation mechanisms and targets for developing antibiofilm agents. Future Med. Chem. 2015, 7, 493–512. [Google Scholar] [CrossRef]
- Deva, A.K.; Adams, W.P.; Vickery, K. The Role of Bacterial Biofilms in Device-Associated Infection. Plast. Reconstr. Surg. 2013, 132, 1319–1328. [Google Scholar] [CrossRef] [Green Version]
- Jamal, M.; Ahmad, W.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.; Rafiq, M.; Kamil, M.A. Bacterial biofilm and associated infections. J. Chin. Med. Assoc. 2018, 81, 7–11. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J. The biofilm matrix. Nat. Rev. Genet. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Aoudia, N.; Rieu, A.; Briandet, R.; Deschamps, J.; Chluba, J.; Jego, G.; Garrido, C.; Guzzo, J. Biofilms of Lactobacillus plantarum and Lactobacillus fermentum: Effect on stress responses, antagonistic effects on pathogen growth and immunomodulatory properties. Food Microbiol. 2016, 53, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Floyd, K.; Eberly, A.; Hadjifrangiskou, M. Adhesion of bacteria to surfaces and biofilm formation on medical devices. In Biofilms and Implantable Medical Devices; Elsevier Inc.: New York, NY, USA, 2017; pp. 47–95. [Google Scholar]
- Maharjan, G.; Khadka, P.; Shilpakar, G.S.; Chapagain, G.; Dhungana, G.R. Catheter-Associated Urinary Tract Infection and Obstinate Biofilm Producers. Can. J. Infect. Dis. Med. Microbiol. 2018, 2018, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tunney, M.M.; Gorman, S.P.; Patrick, S. Infection associated with medical devices. Int. J. Gen. Syst. 2012, 31, 195–205. [Google Scholar] [CrossRef]
- Mandakhalikar, K.D.; Chua, R.R.; Tambyah, P.A. New Technologies for Prevention of Catheter Associated Urinary Tract Infection. Curr. Treat. Options Infect. Dis. 2016, 8, 24–41. [Google Scholar] [CrossRef]
- Barzegari, A.; Kheyrolahzadeh, K.; Khatibi, S.M.H.; Sharifi, S.; Memar, M.Y.; Vahed, S.Z. The Battle of Probiotics and Their Derivatives against Biofilms. Infect. Drug Resist. 2020, 13, 659–672. [Google Scholar] [CrossRef] [Green Version]
- Sambanthamoorthy, K.; Feng, X.; Patel, R.; Patel, S.; Paranavitana, C. Antimicrobial and antibiofilm potential of biosurfactants isolated from lactobacilli against multi-drug-resistant pathogens. BMC Microbiol. 2014, 14, 197. [Google Scholar] [CrossRef] [Green Version]
- Kaur, S.; Sharma, P.; Kalia, N.; Singh, J.; Kaur, S. Faculty Opinions recommendation of Anti-biofilm Properties of the Fecal Probiotic Lactobacilli Against Vibrio spp. Front. Cell. Infect. Microbiol. 2019, 8, 1–14. [Google Scholar]
- Otero, M.C.; Nader-Macías, M. Inhibition of Staphylococcus aureus by H2O2-producing Lactobacillus gasseri isolated from the vaginal tract of cattle. Anim. Reprod. Sci. 2006, 96, 35–46. [Google Scholar] [CrossRef]
- Markowiak, P.; Śliżewska, K.; Markowiak, P.; Śliżewska, K. Effects of Probiotics, Prebiotics, and Synbiotics on Human Health. Nutrients 2017, 9, 1021. [Google Scholar] [CrossRef]
- FAO. Probiotics in Food: Health and Nutritional Properties and Guidelines for Evaluation. 2006. Available online: http://www.fao.org/3/a-a0512e.pdf (accessed on 7 December 2020).
- Piqué, N.; Berlanga, M.; Miñana-Galbis, D. Health Benefits of Heat-Killed (Tyndallized) Probiotics: An Overview. Int. J. Mol. Sci. 2019, 20, 2534. [Google Scholar] [CrossRef] [Green Version]
- Williams, N.T. Probiotics. Am. J. Health Pharm. 2010, 67, 449–458. [Google Scholar] [CrossRef]
- Prabhurajeshwar, C.; Chandrakanth, R.K. Probiotic potential of Lactobacilli with antagonistic activity against pathogenic strains: An in vitro validation for the production of inhibitory substances. Biomed. J. 2017, 40, 270–283. [Google Scholar] [CrossRef] [PubMed]
- de Melo Pereira, G.V.; Coelho, B.D.O.; Júnior, A.I.M.; Thomaz-Soccol, V.; Soccol, C.R. How to select a probiotic? A review and update of methods and criteria. Biotechnol. Adv. 2018, 36, 2060–2076. [Google Scholar] [CrossRef] [PubMed]
- Fioramonti, J.; Theodorou, V.; Bueno, L. Probiotics: What are they? What are their effects on gut physiology? Best Pr. Res. Clin. Gastroenterol. 2003, 17, 711–724. [Google Scholar] [CrossRef]
- Gogineni, V.K.; Morrow, L.E. Probiotics: Mechanisms of Action and Clinical Applications. J. Probiotics Health 2013, 1. [Google Scholar] [CrossRef] [Green Version]
- Ng, S.C.; Hart, A.L.; Kamm, M.A.; Stagg, A.J.; Knight, S.C. Mechanisms of action of probiotics: Recent advances. Inflamm. Bowel Dis. 2009, 15, 300–310. [Google Scholar] [CrossRef]
- Bermudez-Brito, M.; Plaza-Díaz, J.; Muñoz-Quezada, S.; Gómez-Llorente, C.; Gil, A. Probiotic Mechanisms of Action. Ann. Nutr. Metab. 2012, 61, 160–174. [Google Scholar] [CrossRef]
- Salas-Jara, M.J.; Ilabaca, A.; Vega, M.; García-Cancino, A. Biofilm Forming Lactobacillus: New Challenges for the Development of Probiotics. Microorganisms 2016, 4, 35. [Google Scholar] [CrossRef]
- Nemcová, R. Criteria for selection of lactobacilli for probiotic use. Vet. Med. 1997, 42, 19–27. [Google Scholar]
- Oelschlaeger, T.A. Mechanisms of probiotic actions–A review. Int. J. Med. Microbiol. 2010, 300, 57–62. [Google Scholar] [CrossRef]
- Khalighi, A.; Behdani, R.; Kouhestani, S. Probiotics: A Comprehensive Review of Their Classification, Mode of Action and Role in Human Nutrition. In Probiotics and Prebiotics in Human Nutrition and Health; InTech: London, UK, 2016. [Google Scholar]
- Muñoz, M.; Mosquera, A.; Alméciga-Díaz, C.J.; Melendez, A.; Sánchez, O.F. Fructooligosaccharides metabolism and effect on bacteriocin production in Lactobacillus strains isolated from ensiled corn and molasses. Anaerobe 2012, 18, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Kechagia, M.; Basoulis, D.; Konstantopoulou, S.; Dimitriadi, D.; Gyftopoulou, K.; Skarmoutsou, N.; Fakiri, E.M. Health Benefits of Probiotics: A Review. Int. Sch. Res. Not. 2013, 2013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holzapfel, W.; Haberer, P.; Geisen, R.; Björkroth, J.; Schillinger, U. Taxonomy and important features of probiotic microorganisms in food and nutrition. Am. J. Clin. Nutr. 2001, 73, 365s–373s. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carr, F.J.; Chill, D.; Maida, N. The Lactic Acid Bacteria: A Literature Survey. Crit. Rev. Microbiol. 2002, 28, 281–370. [Google Scholar] [CrossRef]
- Leroy, F.; De Vuyst, L. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci. Technol. 2004, 15, 67–78. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; Altman, D.; Antes, G.; Atkins, D.; Barbour, V.; Barrowman, N.; Berlin, J.A.; et al. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009, 6. [Google Scholar] [CrossRef] [Green Version]
- Slim, K.; Nini, E.; Forestier, D.; Kwiatkowski, F.; Panis, Y.; Chipponi, J. Methodological index for non-randomized studies (MINORS): Development and validation of a new instrument. ANZ J. Surg. 2003, 73, 712–716. [Google Scholar] [CrossRef]
- Khan, M.; Rothrauff, B.B.; Merali, F.; Musahl, V.; Peterson, D.; Ayeni, O.R. Management of the Contaminated Anterior Cruciate Ligament Graft. Arthrosc. J. Arthrosc. Relat. Surg. 2014, 30, 236–244. [Google Scholar] [CrossRef]
- Harrer, M.; Cuijpers, P.; Furukawa, T.A.; Ebert, D.D. Doing Meta-Analysis in R: A Hands-on Guide. 2019. Available online: https://bookdown.org/MathiasHarrer/Doing_Meta_Analysis_in_R/ (accessed on 1 September 2020).
- Higgins, J.P.T.; Thompson, S.G.; Deeks, J.J.; Altman, D.G. Measuring inconsistency in meta-analyses. BMJ 2003, 327, 557–560. [Google Scholar] [CrossRef] [Green Version]
- Borenstein, M.; Hedges, L.V.; Higgins, J.P.T.; Rothstein, H.R. Identifying and Quantifying Heterogeneity. In Introduction to Meta-Analysis; John Wiley & Sons: Hoboken, NJ, USA, 2009; pp. 107–125. [Google Scholar]
- Borenstein, M.; Hedges, L.V.; Higgins, J.P.T.; Rothstein, H.R. Publication Bias. In Introduction to Meta-Analysis; John Wiley & Sons: Hoboken, NJ, USA, 2009; pp. 277–292. [Google Scholar]
- Walencka, E.; Różalska, S.; Sadowska, B. The influence of Lactobacillus acidophilus-derived surfactants on staphylococcal adhesion and biofilm formation. Folia Microbiol. 2008, 53, 61–66. [Google Scholar] [CrossRef]
- Ceresa, C.; Tessarolo, F.; Caola, I.; Nollo, G.; Cavallo, M.; Rinaldi, M.; Fracchia, L. Inhibition of Candida albicans adhesion on medical-grade silicone by a Lactobacillus-derived biosurfactant. J. Appl. Microbiol. 2015, 118, 1116–1125. [Google Scholar] [CrossRef]
- Cadieux, P.; Watterson, J.D.; Denstedt, J.; Harbottle, R.R.; Puskas, J.; Howard, J.; Gan, B.S.; Reid, G. Potential application of polyisobutylene-polystyrene and a Lactobacillus protein to reduce the risk of device-associated urinary tract infections. Colloids Surf. B Biointerfaces 2003, 28, 95–105. [Google Scholar] [CrossRef]
- Mohapatra, A.R.; Jeevaratnam, K. Inhibiting bacterial colonization on catheters: Antibacterial and antibiofilm activities of bacteriocins from Lactobacillus plantarum SJ33. J. Glob. Antimicrob. Resist. 2019, 19, 85–92. [Google Scholar] [CrossRef]
- Van Der Mei, H.C.; Van De Belt-Gritter, B.; Van Weissenbruch, R.; Dijk, F.; Albers, F.; Busscher, H. Effect of Consumption of Dairy Products with Probiotic Bacteria on Biofilm Formation on Silicone Rubber Implant Surfaces in an Artificial Throat. Food Bioprod. Process. 1999, 77, 156–158. [Google Scholar] [CrossRef]
- Rodrigues, L.; Van Der Mei, H.; Banat, I.M.; Teixeira, J.; Oliveira, R.; Rodrigues, L.R.; Teixeira, J.A. Inhibition of microbial adhesion to silicone rubber treated with biosurfactant from Streptococcus thermophiles A. FEMS Immunol. Med. Microbiol. 2006, 46, 107–112. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, L.R.; Van Der Mei, H.; Teixeira, J.A.; Oliveira, R. Biosurfactant from Lactococcus lactis 53 inhibits microbial adhesion on silicone rubber. Appl. Microbiol. Biotechnol. 2004, 66, 306–311. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Q.; Stamatova, I.; Kainulainen, V.; Ič, R.K.; Meurman, J.H. Interactions between Lactobacillus rhamnosus GG and oral micro-organisms in an in vitro biofilm model. BMC Microbiol. 2016, 16, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Ciandrini, E.; Campana, R.; Casettari, L.; Perinelli, D.R.; Fagioli, L.; Manti, A.; Bonacucina, G.; Papa, S.; Baffone, W. Characterization of biosurfactants produced by Lactobacillus spp. and their activity against oral streptococci biofilm. Appl. Microbiol. Biotechnol. 2016, 100, 6767–6777. [Google Scholar] [CrossRef]
- Song, Y.-G.; Lee, S.-H. Inhibitory effects of Lactobacillus rhamnosus and Lactobacillus casei on Candida biofilm of denture surface. Arch. Oral Biol. 2017, 76, 1–6. [Google Scholar] [CrossRef]
- Rodrigues, L.R.; Banat, I.M.; Teixeira, J.; Oliveira, R. Biosurfactants: Potential applications in medicine. J. Antimicrob. Chemother. 2006, 57, 609–618. [Google Scholar] [CrossRef]
- Fracchia, L.; Cavallo, M.; Giovanna, M.; Banat, I.M. Biosurfactants and Bioemulsifiers Biomedical and Related Applications–Present Status and Future Potentials. Biomed. Sci. Eng. Technol. 2012, 325–370. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, L.R.; Teixeira, J.A.; Van Der Mei, H.C.; Oliveira, R. Isolation and partial characterization of a biosurfactant produced by Streptococcus thermophilus A. Colloids Surf. B Biointerfaces 2006, 53, 105–112. [Google Scholar] [CrossRef] [Green Version]
- Gudiña, E.J.; Fernandes, E.C.; Teixeira, J.A.; Rodrigues, L.R. Antimicrobial and anti-adhesive activities of cell-bound biosurfactant from Lactobacillus agilis CCUG31450. RSC Adv. 2015, 5, 90960–90968. [Google Scholar] [CrossRef] [Green Version]
- Tahmourespour, A.; Salehi, R.; Kermanshahi, R.; Eslami, G. The anti-biofouling effect of Lactobacillus fermentum-derived biosurfactant against Streptococcus mutans. Biofouling 2011, 27, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Gudiña, E.J.; Teixeira, J.A.; Rodrigues, L.R. Isolation and functional characterization of a biosurfactant produced by Lactobacillus paracasei. Colloids Surf. B Biointerfaces 2010, 76, 298–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tahmourespour, A.; Salehi, R.; Kermanshahi, R.K. Lactobacillus Acidophilus-Derived Biosurfactant Effect on GTFB and GTFC Expression Level in Streptococcus Mutans Biofilm Cells. Braz. J. Microbiol. 2011, 42, 330–339. [Google Scholar] [CrossRef] [Green Version]
- Varjani, S.; Upasani, V.N. Critical review on biosurfactant analysis, purification and characterization using rhamnolipid as a model biosurfactant. Bioresour. Technol. 2017, 232, 389–397. [Google Scholar] [CrossRef]
- Banat, I.M.; Franzetti, A.; Gandolfi, I.; Bestetti, G.; Martinotti, M.G.; Fracchia, L.; Smyth, T.J.; Marchant, R. Microbial biosurfactants production, applications and future potential. Appl. Microbiol. Biotechnol. 2010, 87, 427–444. [Google Scholar] [CrossRef]
- Satpute, S.K.; Kulkarni, G.R.; Banpurkar, A.G.; Banat, I.M.; Mone, N.S.; Patil, R.H.; Cameotra, S.S. Biosurfactant/s from Lactobacilli species: Properties, challenges and potential biomedical applications. J. Basic Microbiol. 2016, 56, 1140–1158. [Google Scholar] [CrossRef]
- Velraeds, M.M.; Van Der Mei, H.C.; Reid, G.; Busscher, H.J. Physicochemical and biochemical characterization of biosurfactants released by Lactobacillus strains. Colloids Surf. B Biointerfaces 1996, 8, 51–61. [Google Scholar] [CrossRef]
- Satpute, S.K.; Banat, I.M.; Dhakephalkar, P.K.; Banpurkar, A.G.; Chopade, B.A. Biosurfactants, bioemulsifiers and exopolysaccharides from marine microorganisms. Biotechnol. Adv. 2010, 28, 436–450. [Google Scholar] [CrossRef] [PubMed]
- Heyd, M.; Kohnert, A.; Tan, T.-H.; Nusser, M.; Kirschhöfer, F.; Brenner-Weiss, G.; Franzreb, M.; Berensmeier, S. Development and trends of biosurfactant analysis and purification using rhamnolipids as an example. Anal. Bioanal. Chem. 2008, 391, 1579–1590. [Google Scholar] [CrossRef] [PubMed]
- Gudiña, E.J.; Rocha, V.; Teixeira, J.A.; Rodrigues, L.R. Antimicrobial and antiadhesive properties of a biosurfactant isolated from Lactobacillus paracasei ssp. paracasei A20. Lett. Appl. Microbiol. 2010, 50, 419–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morais, I.M.C.; Cordeiro, A.L.; Teixeira, G.S.; Domingues, V.S.; Nardi, R.M.D.; Monteiro, A.S.; Alves, R.J.; Siqueira, E.P.; Santos, V.L. Biological and physicochemical properties of biosurfactants produced by Lactobacillus jensenii P6A and Lactobacillus gasseri P65. Microb. Cell Factories 2017, 16, 1–15. [Google Scholar] [CrossRef]
- Sharma, D.; Saharan, B.S. Functional characterization of biomedical potential of biosurfactant produced by Lactobacillus helveticus. Biotechnol. Rep. 2016, 11, 27–35. [Google Scholar] [CrossRef] [Green Version]
- Tan, Y.; Leonhard, M.; Moser, D.; Schneider-Stickler, B. Inhibition activity of Lactobacilli supernatant against fungal-bacterial multispecies biofilms on silicone. Microb. Pathog. 2017, 113, 197–201. [Google Scholar] [CrossRef]
- Yan, X.; Gu, S.; Cui, X.; Shi, Y.; Wen, S.; Chen, H.; Ge, J. Antimicrobial, anti-adhesive and anti-biofilm potential of biosurfactants isolated from Pediococcus acidilactici and Lactobacillus plantarum against Staphylococcus aureus CMCC26003. Microb. Pathog. 2019, 127, 12–20. [Google Scholar] [CrossRef]
- Al-Mathkhury, H.J.F.; Ali, A.S.; Ghafil, J.A. Antagonistic effect of bacteriocin against urinary catheter associated Pseudomonas aeruginosa biofilm. N. Am. J. Med. Sci. 2011, 3, 367–370. [Google Scholar] [CrossRef]
- Shahandashti, R.V.; Kermanshahi, R.K.; Ghadam, P. The inhibitory effect of bacteriocin produced by Lactobacillus acidophilus ATCC 4356 and Lactobacillus plantarum ATCC 8014 on planktonic cells and biofilms of Serratia marcescens. Turk. J. Med. Sci. 2016, 46, 1188–1196. [Google Scholar] [CrossRef]
- Abid, Y.; Casillo, A.; Gharsallah, H.; Joulak, I.; Lanzetta, R.; Corsaro, M.M.; Attia, H.; Azabou, S. Production and structural characterization of exopolysaccharides from newly isolated probiotic lactic acid bacteria. Int. J. Biol. Macromol. 2018, 108, 719–728. [Google Scholar] [CrossRef]
- James, K.M.; Macdonald, K.W.; Chanyi, R.M.; Cadieux, P.A.; Burton, J.P. Inhibition of Candida albicans biofilm formation and modulation of gene expression by probiotic cells and supernatant. J. Med. Microbiol. 2016, 65, 328–336. [Google Scholar] [CrossRef]
- Tan, Y.; Leonhard, M.; Moser, D.; Ma, S.; Schneider-Stickler, B. Inhibitory effect of probiotic lactobacilli supernatants on single and mixed non-albicans Candida species biofilm. Arch. Oral Biol. 2018, 85, 40–45. [Google Scholar] [CrossRef]
- McMillan, A.; Dell, M.; Zellar, M.P.; Cribby, S.; Martz, S.; Hong, E.; Fu, J.; Abbas, A.; Dang, T.; Miller, W.; et al. Disruption of urogenital biofilms by lactobacilli. Colloids Surf. B Biointerfaces 2011, 86, 58–64. [Google Scholar] [CrossRef]
- Rao, K.P.; Chennappa, G.; Suraj, U.; Nagaraja, H.; Raj, A.P.C.; Sreenivasa, M.Y. Probiotic Potential of Lactobacillus Strains Isolated from Sorghum-Based Traditional Fermented Food. Probiotics Antimicrob. Proteins 2015, 7, 146–156. [Google Scholar] [CrossRef]
- Varma, P.; Nisha, N.; Dinesh, K.R.; Kumar, A.V.; Biswas, R. Anti-Infective Properties of Lactobacillus fermentum against Staphylococcus aureus and Pseudomonas aeruginosa. J. Mol. Microbiol. Biotechnol. 2011, 20, 137–143. [Google Scholar] [CrossRef]
- Van Der Mei, H.; Free, R.; Van Weissenbruch, R.; Busscher, H.; Elving, G.; Albers, F.J. Effect of probiotic bacteria on prevalence of yeasts in oropharyngeal biofilms on silicone rubber voice prostheses in vitro. J. Med. Microbiol. 2000, 49, 713–718. [Google Scholar] [CrossRef] [Green Version]
- Song, H.; Zhang, J.; Qu, J.; Liu, J.; Yin, P.; Zhang, G.; Shang, D. Lactobacillus rhamnosus GG microcapsules inhibit Escherichia coli biofilm formation in coculture. Biotechnol. Lett. 2019, 41, 1007–1014. [Google Scholar] [CrossRef]
- Reid, G.; Tieszer, C. Use of lactobacilli to reduce the adhesion of Staphylococcus aureus to catheters. Int. Biodeterior. Biodegrad. 1994, 34, 73–83. [Google Scholar] [CrossRef]
- Dds, C.E.F.; Giacaman, R.A.; Tenuta, L.M.A.; Cury, J.A. Effect of the Probiotic Lactobacillus rhamnosus LB21 on the Cariogenicity of Streptococcus mutans UA159 in a Dual-Species Biofilm Model. Caries Res. 2015, 49, 583–590. [Google Scholar] [CrossRef]
- Ciandrini, E.; Campana, R.; Baffone, W. Live and heat-killed Lactobacillus spp. interfere with Streptococcus mutans and Streptococcus oralis during biofilm development on titanium surface. Arch. Oral Biol. 2017, 78, 48–57. [Google Scholar] [CrossRef]
- Kim, A.R.; Ahn, K.B.; Yun, C.-H.; Park, O.-J.; Perinpanayagam, H.; Yoo, Y.-J.; Kum, K.Y.; Han, S.H. Lactobacillus plantarum Lipoteichoic Acid Inhibits Oral Multispecies Biofilm. J. Endod. 2019, 45, 310–315. [Google Scholar] [CrossRef] [PubMed]
- Ahn, K.B.; Baik, J.E.; Park, O.-J.; Yun, C.-H.; Han, S.H. Lactobacillus plantarum lipoteichoic acid inhibits biofilm formation of Streptococcus mutans. PLoS ONE 2018, 13, e0192694. [Google Scholar] [CrossRef] [Green Version]
- Satpute, S.K.; Mone, N.S.; Das, P.; Banat, I.M.; Banpurkar, A.G. Inhibition of pathogenic bacterial biofilms on PDMS based implants by L. acidophilus derived biosurfactant. BMC Microbiol. 2019, 19, 1–15. [Google Scholar] [CrossRef]
- Gomaa, E.Z. Antimicrobial and anti-adhesive properties of biosurfactant produced by lactobacilli isolates, biofilm formation and aggregation ability. J. Gen. Appl. Microbiol. 2013, 59, 425–436. [Google Scholar] [CrossRef] [Green Version]
- Velraeds, M.M.C.; van de Belt-Gritter, B.; van der Mei, H.C.; Reid, G.; Busscher, H.J. Interference in initial adhesion of uropathogenic bacteria and yeasts to silicone rubber by a Lactobacillus acidophilus biosurfactant. J. Med. Microbiol. 1998, 47, 1081–1085. [Google Scholar] [CrossRef]
- Shokouhfard, M.; Kermanshahi, R.K.; Vahedi-Shahandashti, R.; Feizabadi, M.M.; Teimourian, S. The inhibitory effect of a Lactobacillus acidophilus derived biosurfactant on biofilm producer Serratia marcescens. Iran. J. Basic Med. Sci. 2015, 18, 1001–1007. [Google Scholar]
- Sharma, V.; Harjai, K.; Shukla, G. Effect of bacteriocin and exopolysaccharides isolated from probiotic on P. aeruginosa PAO1 biofilm. Folia Microbiol. 2018, 63, 181–190. [Google Scholar] [CrossRef]
- Comelli, E.M.; Guggenheim, B.; Stingele, F.; Neeser, J.-R. Selection of dairy bacterial strains as probiotics for oral health. Eur. J. Oral Sci. 2002, 110, 218–224. [Google Scholar] [CrossRef]
- Derakhshandeh, S.; Shahrokhi, N.; Khalaj, V.; Habibi, M.; Moazzezy, N.; Karam, M.R.A.; Bouzari, S. Surface display of uropathogenic Escherichia coli FimH in Lactococcus lactis: In vitro characterization of recombinant bacteria and its protectivity in animal model. Microb. Pathog. 2020, 141, 103974. [Google Scholar] [CrossRef]
- Tahmourespour, A.; Kermanshahi, R.K. The effect of a probiotic strain (Lactobacillus acidophilus) on the plaque formation of oral Streptococci. Bosn. J. Basic Med. Sci. 2011, 11, 37–40. [Google Scholar] [CrossRef] [Green Version]
- Ifeoma, M.E.; Jennifer, U.K.; Ezeonu, I.M.; Kanu, J.U. Inhibition of biofilms on urinary catheters using immobilized Lactobacillus cells. Afr. J. Microbiol. Res. 2016, 10, 920–929. [Google Scholar] [CrossRef] [Green Version]
- Sarikaya, H.; Aslim, B.; Yuksekdag, Z.N. Assessment of anti-biofilm activity and bifidogenic growth stimulator (BGS) effect of lyophilized exopolysaccharides (l-EPSs) from Lactobacilli strains. Int. J. Food Prop. 2017, 20, 362–371. [Google Scholar] [CrossRef] [Green Version]
- Chapman, C.M.C.; Gibson, G.R.; Rowland, I. Effects of single- and multi-strain probiotics on biofilm formation and invitro adhesion to bladder cells by urinary tract pathogens. Anaerobe 2014, 27, 71–76. [Google Scholar] [CrossRef]
- Maldonado, N.C.; de Ruiz, C.S.; Cecilia, M.; Nader-Macias, M.E. A simple technique to detect Klebsiella biofilm-forming-strains. Inhibitory potential of Lactobacillus fermentum CRL 1058 whole cells and products. In Communicating Current Research and Educational Topics and Trends in Applied Microbiology; Vilas, A.M., Ed.; FORMATEX: Badajoz, Spain, 2007; pp. 52–59. [Google Scholar]
- Kimelman, H.; Shemesh, M. Probiotic Bifunctionality of Bacillus subtilis-Rescuing Lactic Acid Bacteria from Desiccation and Antagonizing Pathogenic Staphylococcus aureus. Microorganisms 2019, 7, 407. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.C.; Lin, C.T.; Wu, C.Y.; Peng, W.S.; Lee, M.J.; Tsai, Y.C. Inhibitory effect of Lactobacillus salivarius on Streptococcus mutans biofilm formation. Mol. Oral Microbiol. 2015, 30, 16–26. [Google Scholar] [CrossRef]
- Fang, K.; Jin, X.; Hong, S.H. Probiotic Escherichia coli inhibits biofilm formation of pathogenic E. coli via extracellular activity of DegP. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Hancock, V.; Dahl, M.; Klemm, P. Probiotic Escherichia coli strain Nissle 1917 outcompetes intestinal pathogens during biofilm formation. J. Med. Microbiol. 2010, 59, 392–399. [Google Scholar] [CrossRef] [Green Version]
- Benmechernene, Z.; Fernandez-No, I.; Kihal, M.; Böhme, K.; Calo-Mata, P.; Barros-Velázquez, J. Recent patents on bacteriocins: Food and biomedical applications. Recent Pat. DNA Gene Seq. 2013, 7, 66–73. [Google Scholar] [CrossRef]
- Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocins—A viable alternative to antibiotics? Nat. Rev. Genet. 2013, 11, 95–105. [Google Scholar] [CrossRef]
- Oh, S.; Kim, S.; Worobo, R.W. Characterization and Purification of a Bacteriocin Produced by a Potential Probiotic Culture, Lactobacillus acidophilus 30SC. J. Dairy Sci. 2000, 83, 2747–2752. [Google Scholar] [CrossRef]
- Okuda, K.-I.; Zendo, T.; Sugimoto, S.; Iwase, T.; Tajima, A.; Yamada, S.; Sonomoto, K.; Mizunoe, Y. Effects of Bacteriocins on Methicillin-Resistant Staphylococcus aureus Biofilm. Antimicrob. Agents Chemother. 2013, 57, 5572–5579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos, V.; Drummond, R.N.; Dias-Souza, M. Bacteriocins as Antimicrobial and Antibiofilm Agents. In Current Developments in Biotechnology and Bioengineering; Elsevier Inc.: New York, NY, USA, 2017; pp. 403–436. [Google Scholar]
- Meade, M.E.; Slattery, A.; Garvey, M. Bacteriocins, Potent Antimicrobial Peptides and the Fight against Multi Drug Resistant Species: Resistance Is Futile? Antibiotics 2020, 9, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soltani, S.; Hammami, R.; Cotter, P.D.; Rebuffat, S.; Said, L.B.; Gaudreau, H.; Bédard, F.; Biron, E.; Drider, D.; Fliss, I. Bacteriocins as a new generation of antimicrobials: Toxicity aspects and regulations. FEMS Microbiol. Rev. 2020. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhao, X.; Yang, Y.; Zhao, A.; Yang, Z. Characterization and bioactivities of an exopolysaccharide produced by Lactobacillus plantarum YW32. Int. J. Biol. Macromol. 2015, 74, 119–126. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, Z.; Qiu, L.; Zhang, F.; Xu, X.; Wei, H.; Tao, X. Characterization and bioactivities of the exopolysaccharide from a probiotic strain of Lactobacillus plantarum WLPL04. J. Dairy Sci. 2017, 100, 6895–6905. [Google Scholar] [CrossRef]
- Mahdhi, A.; Leban, N.; Chakroun, I.; Bayar, S.; Mahdouani, K.; Majdoub, H.; Kouidhi, B. Use of extracellular polysaccharides, secreted by Lactobacillus plantarum and Bacillus spp., as reducing indole production agents to control biofilm formation and efflux pumps inhibitor in Escherichia coli. Microb. Pathog. 2018, 125, 448–453. [Google Scholar] [CrossRef]
- Kim, Y.; Oh, S.; Kim, S.H. Released exopolysaccharide (r-EPS) produced from probiotic bacteria reduce biofilm formation of enterohemorrhagic Escherichia coli O157:H7. Biochem. Biophys. Res. Commun. 2009, 379, 324–329. [Google Scholar] [CrossRef]
- Velraeds, M.M.; Van De Belt-Gritter, B.; Busscher, H.J.; Reid, G.; Van Der Mei, H.C. Inhibition of uropathogenic biofilm growth on silicone rubber in human urine by lactobacilli—A teleologic approach. World J. Urol. 2000, 18, 422–426. [Google Scholar] [CrossRef]
- Azeredo, J.; Azevedo, N.F.; Briandet, R.; Cerca, N.; Coenye, T.; Costa, A.R.; Desvaux, M.; Di Bonaventura, G.; Hébraud, M.; Jaglic, Z.; et al. Critical review on biofilm methods. Crit. Rev. Microbiol. 2017, 43, 313–351. [Google Scholar] [CrossRef] [Green Version]
- Gomes, M.; Gomes, L.C.; Santos, R.T.; Mergulhão, F.J. PDMS in Urinary Tract Devices: Applications, Problems and Potential Solutions. In Polydimethylsiloxane: Structure and Applications; Carlsen, P.N., Ed.; Nova Science Publishers: Hauppauge, NY, USA, 2020. [Google Scholar]
- Köseoğlu, H.; Aslan, G.; Esen, N.; Sen, B.H.; Coban, H. Ultrastructural stages of biofilm development of Escherichia coli on urethral catheters and effects of antibiotics on biofilm formation. Urology 2006, 68, 942–946. [Google Scholar] [CrossRef]
- Stepanović, S.; Ćirković, I.; Mijač, V.; Švabić-Vlahović, M. Influence of the incubation temperature, atmosphere and dynamic conditions on biofilm formation by Salmonella spp. Food Microbiol. 2003, 20, 339–343. [Google Scholar] [CrossRef]
- Stepanović, S.; Vuković, D.; Ježek, P.; Pavlović, M.; Švabic-Vlahović, M. Influence of dynamic conditions on biofilm formation by staphylococci. Eur. J. Clin. Microbiol. Infect. Dis. 2001, 20, 502–504. [Google Scholar] [CrossRef] [PubMed]
- Gänzle, M.G. Lactic metabolism revisited: Metabolism of lactic acid bacteria in food fermentations and food spoilage. Curr. Opin. Food Sci. 2015, 2, 106–117. [Google Scholar] [CrossRef]







| Criterion | Mean |
| 1. A clearly stated aim: The hypothesis/aim of the study is explicitly stated and testable by statistical means. | 1.93 |
| 2. Detection of bias: Data were collected according to an established protocol. At least 3 independent experiments were performed for each assay. | 1.64 |
| 3. An adequate control group: There is a control group corresponding to untreated biofilms. | 1.93 |
| 4. Appropriate methodology: Description and explanation of the methods in accordance with the desired outcomes. The used methods are the same for control and exposure treatment. | 1.84 |
| 5. Pathogens description: The pathogens species and quantity used for inoculation are described. 0: not reported 1: organism species OR organism quantity 2: organism species AND organism quantity | 1.64 |
| 6. Antibiofilm substances: Description of substances used to control/prevent biofilm formation, including identity/origin and concentration. 0: not reported 1: description of origin OR concentration 2: description of origin AND concentration | 1.87 |
| 7. Culture conditions: Description of how assays were performed in sufficient detail to repeat (or detailed methodology is referenced), including culture medium, hydrodynamic conditions and temperature. 0: not described 1: sufficient detail to repeat OR a description of culture medium OR hydrodynamic conditions OR temperature 2: sufficient detail to repeat AND a description of culture medium AND hydrodynamic conditions AND temperature | 1.27 |
| 8. Biofilm formation period: Because some microorganisms may grow/act slower, longer incubation periods may be needed to ensure successful biofilm inhibition. 0: duration of exposure not reported 1: culture of < 6 h 2: culture of ≥ 6 h | 1.76 |
| 9. Surface: Description of substratum for biofilm formation. 0: not described 1: description of surface OR biofilm platform 2: description of surface AND biofilm platform | 1.84 |
| 10. Predictive value: In vitro studies may use inoculum concentrations exceeding those encountered in a clinical scenario. 0: not described 1: inoculation with flora at the same concentration as that found in clinical scenario (<105 CFU/mL) 2: inoculation with a concentration of bacteria that exceeds that found in clinical scenario (>105 CFU/mL) | 1.07 |
| 11. Results clarity: The results of the study are presented in a clear and organized way. 0: results are not clear 1: results are clear 2: results are clear and easy to understand AND cell concentrations or optical density values either for control or treatment experiments were reported | 1.58 |
| 12. Adequate statistical analyses: Description and implementation of statistical tests appropriate to the dataset, with the calculation of confidence intervals and p values. | 1.31 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carvalho, F.M.; Teixeira-Santos, R.; Mergulhão, F.J.M.; Gomes, L.C. The Use of Probiotics to Fight Biofilms in Medical Devices: A Systematic Review and Meta-Analysis. Microorganisms 2021, 9, 27. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms9010027
Carvalho FM, Teixeira-Santos R, Mergulhão FJM, Gomes LC. The Use of Probiotics to Fight Biofilms in Medical Devices: A Systematic Review and Meta-Analysis. Microorganisms. 2021; 9(1):27. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms9010027
Chicago/Turabian StyleCarvalho, Fábio M., Rita Teixeira-Santos, Filipe J. M. Mergulhão, and Luciana C. Gomes. 2021. "The Use of Probiotics to Fight Biofilms in Medical Devices: A Systematic Review and Meta-Analysis" Microorganisms 9, no. 1: 27. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms9010027