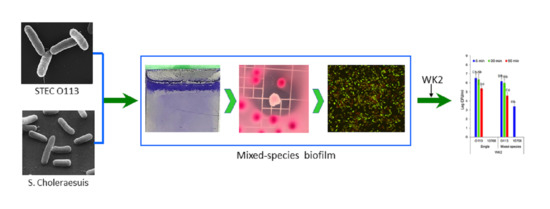
Single- and Dual-Species Biofilm Formation by Shiga Toxin-Producing Escherichia coli and Salmonella, and Their Susceptibility to an Engineered Peptide WK2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Cultivation
2.2. Food Contact Surface Materials and Preparation of Beef Juice
2.3. Preparation of Single- and Mixed-Species Cultures
2.4. Single- and Mixed-Species Biofilm Formation on Stainless Steel Surface in Beef Juice
2.5. Enumeration of the Planktonic and Attached Cells
2.6. Peptide Synthesis
2.7. Peptide Testing
2.8. Sensitivity of STEC O113:H21 and S. Choleraesuis ATCC 10708 in Single- and Mixed-Species Biofilms to WK2
2.9. Confocal Laser Scanning Microscopy (CLSM)
2.10. Statistical Analysis
3. Results
3.1. Single- and Mixed-Species Biofilm Mass Measured by CV Staining
3.2. Enumeration of the Single- and Mixed-Species Planktonic and Attached Bacterial Cells
3.3. The Susceptibility of S. Choleraesuis 10708 and STEC O113:H21 in Single- and Mixed-Species Biofilms to WK2
3.4. Microscopic Examination of Single- and Mixed-Species Biofilms
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Byrne, L.; Adams, N.; Jenkins, C. Association between Shiga toxin-producing Escherichia coli O157: H7 stx gene subtype and disease severity, Rngland, 2009–2019. Emerg. Infect. Dis. 2020, 26, 2394–2400. [Google Scholar] [CrossRef]
- Wang, F.; Yang, Q.; Kase, J.A.; Meng, J.; Clotilde, L.M.; Lin, A.; Ge, B. Current trends in detecting non-O157 Shiga toxin-producing Escherichia coli in food. Foodborne Pathog. Dis. 2013, 10, 665–677. [Google Scholar] [CrossRef]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.-A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the united states—major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Giaouris, E.; Simões, M.; Dubois-Brissonnet, F. The role of biofilms in the development and dissemination of microbial resistance within the food industry. Foods 2020, 9, 816. [Google Scholar] [CrossRef]
- Almeida, C.; Azevedo, N.F.; Santos, S.; Keevil, C.W.; Vieira, M.J. Discriminating multi-species populations in biofilms with peptide nucleic acid fluorescence in situ hybridization (pna fish). PLoS ONE 2011, 6, e14786. [Google Scholar] [CrossRef] [Green Version]
- Burmølle, M.; Ren, D.; Bjarnsholt, T.; Sørensen, S.J. Interactions in multispecies biofilms: Do they actually matter? Trends Microbiol. 2014, 22, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Kalchayanand, N.; Brichta-Harhay, D.M.; Arthur, T.M.; Bosilevac, J.M.; Guerini, M.N.; Wheeler, T.L.; Shackelford, S.D.; Koohmaraie, M. Prevalence rates of Escherichia coli O157: H7 and Salmonella at different sampling sites on cattle hides at a feedlot and processing plant. J. Food Prot. 2009, 72, 1267–1271. [Google Scholar] [CrossRef] [Green Version]
- Stephens, T.; Loneragan, G.; Karunasena, E.; Brashears, M. Reduction of Escherichia coli O157 and Salmonella in feces and on hides of feedlot cattle using various doses of a direct-fed microbial. J. Food Prot. 2007, 70, 2386–2391. [Google Scholar] [CrossRef] [PubMed]
- Callejón, R.M.; Rodriguez-Naranjo, M.I.; Ubeda, C.; Hornedo-Ortega, R.; Garcia-Parrilla, M.C.; Troncoso, A.M. Reported foodborne outbreaks due to fresh produce in the United States and European Union: Trends and causes. Foodborne Pathog. Dis. 2015, 12, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Jahid, I.K.; Ha, S.-D. A review of microbial biofilms of produce: Future challenge to food safety. Food Sci. Biotechnol. 2012, 21, 299–316. [Google Scholar] [CrossRef]
- Uhlich, G.A.; Rogers, D.P.; Mosier, D.A. Escherichia coli serotype O157: H7 retention on solid surfaces and peroxide resistance is enhanced by dual-strain biofilm formation. Foodborne Pathog. Dis. 2010, 7, 935–943. [Google Scholar] [CrossRef]
- Chen, D.; Zhao, T.; Doyle, M.P. Single-and mixed-species biofilm formation by Escherichia coli coli O157: H7 and Salmonella, and their sensitivity to levulinic acid plus sodium dodecyl sulfate. Food Control 2015, 57, 48–53. [Google Scholar] [CrossRef]
- Smet, C.; Govaert, M.; Kyrylenko, A.; Easdani, M.; Walsh, J.L.; Van Impe, J.F. Inactivation of single strains of Listeria monocytogenes and Salmonella Typhimurium planktonic cells biofilms with plasma activated liquids. Front Microbiol. 2019, 10, 1539. [Google Scholar] [CrossRef]
- Pletzer, D.; Hancock, R.E. Antibiofilm peptides: Potential as broad-spectrum agents. J. Bacteriol. 2016, 198, 2572–2578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, L.; Hansen, M.F.; Røder, H.L.; Wang, N.; Burmølle, M.; He, G. Mixed-species biofilms in the food industry: Current knowledge and novel control strategies. Crit. Rev. Food Sci. Nutr. 2020, 60, 2277–2293. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Zhang, R.; Hai, D.; Lu, Z.; Lv, F.; Zhao, H.; Zhang, C.; McAllister, T.A.; Stanford, K.; Bie, X. Antibiofilm activity and modes of action of a novel β-sheet peptide against multidrug-resistant Salmonella enterica. Food Res. Int. 2019, 125, 108520. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Stanford, K.; Bie, X.M.; Niu, Y.D.; McAllister, T.A. Effects of beef juice on biofilm formation by Shiga toxin-producing Escherichia coli on stainless steel. Foodborne Pathog. Dis. 2019, 17, 235–242. [Google Scholar] [CrossRef]
- Kumar, A.; Alam, A.; Rani, M.; Ehtesham, N.Z.; Hasnain, S.E. Biofilms: Survival and defense strategy for pathogens. Int. J. Med. Microbiol. 2017, 307, 481–489. [Google Scholar] [CrossRef] [PubMed]
- Abebe, G.M. The role of bacterial biofilm in antibiotic resistance and food contamination. Int. J. Microbiol. 2020, 2020, 1705814. [Google Scholar] [CrossRef]
- Wagner, E.M.; Pracser, N.; Thalguter, S.; Fischel, K.; Rychli, K. Identification of biofilm hotspots in a meat processing environment: Detection of spoilage bacteria in multi-species biofilms. Int. J. Food Microbiol. 2020, 328, 108668. [Google Scholar] [CrossRef]
- Lim, E.S.; Lee, J.E.; Kim, J.S.; Koo, O.K. Isolation of indigenous bacteria from a cafeteria kitchen and their biofilm formation and disinfectant susceptibility. LWT 2017, 77, 376–382. [Google Scholar] [CrossRef]
- Sadekuzzaman, M.; Yang, S.; Mizan, M.F.; Ha, S.D. Current and recent advanced strategies for combating biofilms. Compr. Rev. Food Sci. Food Saf. 2015, 14, 491–509. [Google Scholar] [CrossRef]
- Wang, J.; Stanford, K.; McAllister, T.A.; Johnson, R.P.; Chen, J.; Hou, H.; Zhang, G.; Niu, Y.D. Biofilm formation, virulence gene profiles, and antimicrobial resistance of nine serogroups of non-O157 Shiga toxin-producing Escherichia coli. Foodborne Pathog. Dis. 2016, 13, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Walker, S.L.; Sojka, M.; Dibb-Fuller, M.; Woodward, M.J. Effect of pH, temperature and surface contact on the elaboration of fimbriae and flagella by Salmonella serotype enteritidis. J. Med. Microbiol. 1999, 48, 253–261. [Google Scholar] [CrossRef]
- Mitri, S.; Foster, K.R. The genotypic view of social interactions in microbial communities. Annu. Rev. Genet. 2013, 47, 247–273. [Google Scholar] [CrossRef] [PubMed]
- Lories, B.; Roberfroid, S.; Dieltjens, L.; De Coster, D.; Foster, K.R.; Steenackers, H.P. Biofilm bacteria use stress responses to detect and respond to competitors. Curr. Biol. 2020, 30, 1231–1244. [Google Scholar] [CrossRef] [PubMed]
- Reisner, A.; Höller, B.M.; Molin, S.; Zechner, E.L. Synergistic effects in mixed Escherichia coli biofilms: Conjugative plasmid transfer drives biofilm expansion. J. Bacteriol. 2006, 188, 3582–3588. [Google Scholar] [CrossRef] [Green Version]
- Madsen, J.S.; Burmølle, M.; Hansen, L.H.; Sørensen, S.J. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immuno Med. Microbiol. 2012, 65, 183–195. [Google Scholar] [CrossRef] [Green Version]
- Lappin-Scott, H.M.; Bass, C. Biofilm formation: Attachment, growth, and detachment of microbes from surfaces. Am. J. Infect. Control 2001, 29, 250–251. [Google Scholar] [CrossRef]
- Wang, R.; Kalchayanand, N.; Schmidt, J.W.; Harhay, D.M. Mixed biofilm formation by Shiga toxin-producing Escherichia coli and Salmonella enterica serovar Typhimurium enhanced bacterial resistance to sanitization due to extracellular polymeric substances. J. Food Prot. 2013, 76, 1513–1522. [Google Scholar] [CrossRef] [Green Version]
- Silva, P.L.; Goulart, L.R.; Reis, T.F.; Mendonça, E.P.; Melo, R.T.; Penha, V.A.; Peres, P.A.; Hoepers, P.G.; Beletti, M.E.; Fonseca, B.B. Biofilm formation in different Salmonella serotypes isolated from poultry. Curr. Microbiol. 2019, 76, 124–129. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Z.; Tang, X.; Stanford, K.; Chen, X.; McAllister, T.A.; Niu, Y.D. Single- and Dual-Species Biofilm Formation by Shiga Toxin-Producing Escherichia coli and Salmonella, and Their Susceptibility to an Engineered Peptide WK2. Microorganisms 2021, 9, 2510. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms9122510
Ma Z, Tang X, Stanford K, Chen X, McAllister TA, Niu YD. Single- and Dual-Species Biofilm Formation by Shiga Toxin-Producing Escherichia coli and Salmonella, and Their Susceptibility to an Engineered Peptide WK2. Microorganisms. 2021; 9(12):2510. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms9122510
Chicago/Turabian StyleMa, Zhi, Xia Tang, Kim Stanford, Xiaolong Chen, Tim A. McAllister, and Yan D. Niu. 2021. "Single- and Dual-Species Biofilm Formation by Shiga Toxin-Producing Escherichia coli and Salmonella, and Their Susceptibility to an Engineered Peptide WK2" Microorganisms 9, no. 12: 2510. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms9122510