2. Materials and Methods
This study presents an experimental protocol used to evaluate and assess different types of GANs for augmenting GM datasets. Through experimenting with different architectures, configurations and training strategies, this study aims to propose an optimal architecture for augmenting data of this type. In order to evaluate these results, both descriptive statistics and equivalency testing have been used.
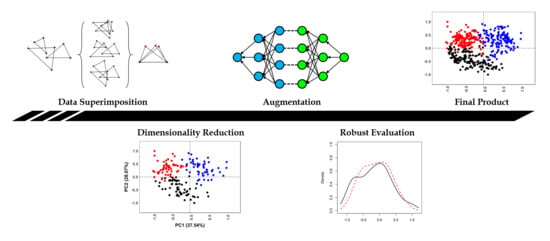
Figure 2 presents a general visualization of the described workflow.
2.1. Datasets
Experiments included within this study were performed on a total of three GM datasets. These datasets originated from experimental archaeology samples in taphonomy. Each of these samples thus represent the morphological features of microscopic alterations observed on bone, using GM to quantify these morphologies for diagnostic purposes.
Nevertheless, considering the objective of this study is to observe the effects of generative learning for GM data augmentation, the origin of these datasets was considered unimportant. The reason behind this lies in how, regardless of the element under study, data used for GM analysis consists of superimposed landmark coordinates (
Figure 1 and
Figure 2). From these coordinates, dimensionality reduction can be used to convert each element into a single vector from which models can learn from (
Figure 2). Therefore, irrespective of whether the raw landmark data was obtained from paleoanthropological specimens, lithic tools or carnivore feeding samples (
Figure 1), all landmarks are similarly embedded into a single vector that can be used as the input to our computational models. Additional use of these three case studies was based on how each dataset was personally generated by the corresponding author, providing a means of controlling the origin of information.
The 3 datasets used consists of a mixture of manually placed landmarks (Type II or III;
Figure 1), as well as some computational semi-landmarks [
3,
4]. The three datasets include;
Dataset 1 (DS1); canid tooth score dataset [
16]. This dataset consists of 105 individuals from three different experimental carnivore feeding samples (labelled foxes, dogs and wolves). 3D models for data extraction were generated using a low-cost structured light surface scanner (David SLS-2). The topography of each 3D digital model was then used to extract 2D images where landmarks could be placed. Landmark data consist of a mixture of Type II and Type III 2D landmarks.
Dataset 2 (DS2); scratch and graze trampling dataset [
15]. This dataset consists of 60 individuals from two different experimental trampling mark samples (labelled scratches and grazes). Each of the elements under study were digitized employing a 3D Digital Microscope (HIROX KH-8700), using between 100× and 200× magnification. Collection of landmark data was then performed following a series of measurements that established a 3D coordinate system across the model. Landmark data consist of a mixture of Type II and Type III 3D landmarks.
Dataset 3 (DS3); semi-landmark based tooth pit dataset [
29]. This dataset consists of an adaptation of DS1 using 60 individuals from two carnivore feeding samples (labelled dogs and wolves). 3D models for data extraction were generated using a low-cost structured light surface scanner (David SLS-2). Landmark data consist of a mixture of 3D Type II landmarks and a mesh of semi-landmarks.
These three datasets were chosen considering the dimensionality of the corresponding feature-space produced for GM analysis (ℝ14, ℝ39 and ℝ60 respectively). With each of these datasets presenting different dimensionalities, optimal GAN architectures could therefore be proposed so as to establish a standardized protocol, regardless of the target domain’s ℝn size.
These datasets were also chosen to observe the effect original sample size has on the accuracy of synthetic data. The latter was tested via minimum sample size calculations according to Cohen’s
d (power = 0.8, d = 0.8, α = 0.05, ratio = 1:1) [
35]. This established a minimum sample size for two-sample statistical comparisons of 26 individuals, rounded up to 30 for simplicity. In accordance with this calculation, experiments were performed by randomly sampling 30 real individuals and comparing them with 30 synthetic individuals. In datasets where larger samples were available, 60 real individuals were sampled and compared with 60 synthetic data points.
2.2. Baseline Geometric Morphometric Data Acquisition
Each of the datasets were prepared using traditional GM techniques, first performing a full Procrustes fit of landmark coordinates via GPA (
Figure 2), followed by the extraction of multivariate features through PCA [
7,
8]. Considering how the objectives of this study are to find the optimal algorithm for mapping out multidimensional distributions, differences in
shape-size relationships were considered irrelevant for this study. GPA was therefore only performed using fully superimposed coordinates in shape feature space.
From here, PC scores were analyzed evaluating their dimensionality and the proportion of variance represented across each of the decomposed eigenvectors and their eigenvalues. Considering how the final eigenvalues begin to represent little or no variance within the landmark configuration, preference was given to those PC scores representing up to 95% of sample variance for statistical evaluations.
For the purpose of this study, GM pre-processing of samples was performed in the free statistical software R (
https://www.r-project.org/, v.3.5.1 64-bit).
2.3. Generative Adversarial Networks
A GAN is a Deep Learning (DL) architecture used for the synthesis of data via a generator model. GANs are fit to data using an unsupervised approach, where the generator is trained by competing with a discriminator that evaluates the authenticity of the synthetic data produced [
24,
41]. While the basic concept behind a GAN is relatively straightforward, the theory behind their configuration and training can be incredibly challenging [
25,
48,
49,
50,
51].
To generate new data, the generator samples from a random Gaussian distribution (e.g., µ = 0, σ = 1), finding the best means of mapping this data out onto the real sample domain. A fixed-length random vector is used as input, triggering the generative process. Once trained, this vector space can essentially be considered a compressed representation of the real data’s distribution. This multidimensional vector space is most commonly referred to in DL literature as
latent space [
21,
48].
The discriminator model takes as input the output of the generator. This discriminator can then be used to predict a class label (real or fake) for the generated data. In some cases, this model is referred to as a
critic model [
52,
53].
For the purpose of this study, multiple experiments were performed to define an optimal GAN architecture. These experiments followed standard DL protocol, finding the optimal neural network configurations by evaluating the effects of each hyperparameter on model performance. Summaries of the hyperparameters tested are included in
Table 1.
In addition to this, the extensive literature on the “best-practices” in GAN research and different heuristics in GAN hyperparameter selection were considered [
48,
49,
51,
54]. Among these, common “GAN-Hacks” were evaluated, including:
Use of the Adam optimization algorithm (α = 0.0002, β1 = 0.5)
Use of dropout in the generator with a probability threshold of 0.3
Use of Leaky ReLU (slope = 0.2)
Stack hidden layers with increasing size in the generator and decreasing size in the discriminator.
For training, trials experimenting with the number of epochs and batch sizes were performed. The final values were chosen in accordance with the requirements of the model in order to reach an acceptable stability.
While binary cross-entropy is typically a recommended loss function for training, this study experimented with alternatives, such as the Least Squares loss function (
LSGAN) and two versions of Wasserstein loss (
WGAN).
LSGAN was originally proposed as a means of overcoming small or vanishing gradients, which are frequently observed when using binary cross-entropy [
50,
52]. In
LSGAN, the discriminator (
D) attempts to minimize the loss (
L), using the sum squared difference between the predicted and expected values for real and fake data (Equation (1)), while the generator (
G) attempts to minimize this difference assuming data is real (Equation (2)):
This results in a greater penalization of larger errors (E) which forces the model to update weights more frequently, therefore avoiding vanishing gradients [
55].
WGAN, on the other hand, is based on the theory of
Earth-Mover’s distance [
52], calculating the distance between the two probability distributions so that one distribution can be converted into another (Equations (3) and (4)):
WGAN additionally uses weight constraints (hypercube of [−0.01, 0.01]) to ensure that the discriminator lies within a 1-Lipschitz function. In certain cases, however, this has been reported to produce some undesired effects [
53]. As an alternative, a proposed adaptation, in the form of
gradient penalty WGAN (
WGAN-GP), includes the same loss for the generator (Equation (4)) but a modified discriminator (eg., 5) with no weight constraints [
53,
56]:
For both loss functions to work, the output of
D requires a linear activation function. Finally, optimization tests were performed using Adam (α = 0.0002, β
1 = 0.5) and RMSprop (α = 0.00005) [
50,
53,
57,
58].
More details on the mathematical components of each GAN loss function can be consulted in the corresponding references [
25,
50,
52,
53,
55].
GANs were trained on scaled PCA feature spaces with 64-bit values ranging between 1 and −1. This scaling procedure was performed to boost neural network performance and optimization by helping reduce the size of weight updates [
24]. For these experiments, GANs were trained on all data within the dataset, regardless of label. This approach was chosen to directly observe how GANs handle this type of input data before considering more complex applications, including sample labels. (See
Section 2.4)
All experiments were performed in the Python programming language (
https://www.python.org/, v.3.7 64-bit) using TensorFlow (
https://www.tensorflow.org/, v.2.0). Neural networks were compiled and trained on the CPU of an ASUS X550VX laptop (Intel
® Core
TM i5 6300HQ).
2.4. Conditional Generative Adversarial Networks
The final GAN trials performed adapted the optimally defined model in
Section 3.1 for Conditional GAN tasks (CGAN). A CGAN is an extension of traditional GANs that incorporate class labels into the input, thus conditioning the generation process. Class labels are encoded and used as input alongside both the latent vector and the original vector in order for the GAN to learn targeted distributions within the dataset [
59]. This can be done by using an embedding layer and concatenating the embedded information with the original input [
60]. It is recommended that the embedding layer is kept as small as possible [
51], with some of the original implementations of CGANs using an embedding layer with a size of only ≈5% of the original flattened generator’s output. Because 5% of our largest dataset would still have been <1, experiments were performed with different sized embedding layers to find the optimal configuration. The best results came out using a ⌈1/4 •
n⌉ sized embedding layer, where n corresponds to the number of dimensions in ℝ
n for each of the targeted feature spaces.
For comparison of GAN and CGAN performance, these models were used separately to augment the DS3. This dataset was chosen considering it was the most complex feature space to map, the most balanced (when compared with DS1), and the most difficult to study (seeing how DS2 presents the highest natural separability).
2.5. Synthetic Data Evaluation
Evaluation of GANs is a complex issue with little general agreement on suitable evaluation metrics [
49]. Considering how most practitioners in GAN research work with computer vision applications, many papers use manual inspection of images to evaluate synthesized data [
61]. Image evaluation, for example, often consists in the visualization of GAN outputs to check whether they are realistic or not, or the use of specified algorithms that are very image-specific [
61]. For the synthesis of numeric data, manual inspection is evidently a very subjective means of evaluating information, while calculations such as Inception-score are not applicable to this type of data [
49,
61,
62]. Under this premise, the majority of metrics used in GAN literature is of little value to the present study, as they almost exclusively focus on the evaluation of images [
60,
62].
Multidimensional numbers are incredibly difficult to visualize, meaning that precise human inspection of this data is impossible. To overcome this, a number of statistical metrics were adopted for GAN evaluation.
Firstly homogeneity of GM data was tested. In most traditional cases, the elimination of size and preservation of allometry in GPA is known to normalize data [
63]. Nevertheless, this assumption does not always hold true. The first logical step was to therefore evaluate distribution homogeneity and normality via multiple Shapiro tests. Synthetic distributions were then compared with the real data to assess the magnitude of differences and the significance of overlapping. For this, a “Two One-Sided” equivalency Test (TOST) was performed. TOST evaluates the magnitude of similarities between samples by using upper (εS) and lower (εI) equivalence bounds that can be established via Cohen’s
d. This assesses H
0 and H
a using an α threshold of
p < 0.05, with H
a implicating significant similarities among samples [
35,
64,
65,
66,
67]. For TOST the test statistics used to assess these similarities were dependent on distribution normality. These varied between the traditional parametric method using Welch’s
t-statistic [
68], or a trimmed non-parametric approach using Yuen’s robust
t-statistic [
69,
70]. To differentiate between the two, from this point onwards non-parametric robust TOST will be referred to as rTOST.
More traditional univariate descriptive statistics were also employed. For distributions matching Gaussian properties, sample means and standard deviations were calculated. These were accompanied by calculations of sample skewness and kurtosis. For significantly non-Gaussian distributions, robust statistical metrics were used instead. In these cases, measurements of central tendency were established using the sample median (
m), while deviations were calculated using the square root of the Biweight Midvariance (
BWMV) (Equations (6)–(9)) [
29,
71,
72].
Robust skewness and kurtosis values were calculated using trimmed distributions. Trims were established using Interquatile Ranges (IR) [
71], with confidence intervals of
p = [0.05:0.95]. Both the IR range and the trimmed skewness and kurtosis values were reported.
Finally, wherever possible, correlations were calculated to compare the effect of hyperparameters on the quality of synthesized data. For homogeneous data, the parametric Pearson test was used [
73], whereas inhomogeneous data was tested using the non-parametric Kendall τ rank-based test [
74]. Considering neural networks are stochastic in nature, these correlations were performed using data obtained from multiple training runs of each GAN to ensure a more robust calculation.
3. Results
All three datasets analyzed present highly inhomogeneous multivariate distributions (
p < 2.2 × 10
−16). Univariate comparisons (
Table 2), however, present a mixture of both inhomogeneous and homogeneous distributions across PC1 and PC2, where the majority of variance is represented.
GAN failure through mode collapse was frequently observed throughout most of the initial trials, characterized by an intense clustering of points with little to no variation in feature space (
Figure 3). Qualitatively, this type of failure is easily diagnosed by visual inspection of graphs. Quantitatively, mode failure can be characterized by a dramatic decrease in variance seen through deviation metrics. To provide an example,
Figure 1 presents the use of a vanilla GAN trained on DS2. At first, training can be seen to start well with the closest (yet not optimal) approximation to the target domain’s median (
Figure 3A). Nevertheless, little variation is present (
BWMV of PC1 = 0.14, target
BWMV = 0.38). As training continues, the algorithm is unable to find the correct median, and performance is seen to deteriorate (
Figure 3B). This presents an exponential decrease in the variance of synthetic data (
Figure 3B PC1
BWMV = 0.02,
Figure 3C PC1
BWMV = 0.0002). Likewise, through mode collapse, the generator is unable to map the true normality of the distribution, generating increasingly normal data in PC1 (Shapiro
w > 0.98,
p > 0.56).
Replacing Leaky ReLU with tanh activation functions resulted in a significant improvement of generated sample medians (difference in median for Leaky ReLU = 0.78; tanh = 0.26), yet with little improvement in BWMV.
To overcome mode collapse, kernel initializers and batch normalization algorithms were incorporated into both the generator and the discriminator. Batch normalization was included before activation, presenting an increase in BWMV. Initializers required careful adjustment, with small standard deviation values resulting in mode collapse. Additional experiments found the discriminator to require a more intense initializer (σ = 0.1) than the generator (σ = 0.7), while optimal results were obtained using a random normal distribution. Such a configuration allows the generator more room to adjust its weights, finding the best way of reaching the target domain’s median and absolute deviation while preventing the discriminator from learning too quickly.
Experiments adjusting hidden layer densities found symmetry between the generator and the discriminator to be unnecessary. The generator was seen to require more hidden layers in order to learn the distributions efficiently, while a larger density than the output in the last hidden layer also produced an increase in performance. The discriminator worked best with just two hidden layers.
3.1. Optimal Architectures
To optimally adjust these finds with all three datasets, the best GAN architecture that presented no mode collapse was obtained using 3 hidden layers in the generator and two hidden layers in the discriminator. The size of hidden layers are conditioned by the size of the target feature space (
Figure 4). To use the example of the largest target domain (DS3 = ℝ
60), the generator is programmed so that the first hidden layer (
Gh1) is a quarter of the size of the target vector (in this case
Gh1 = 60·1/4= 15). If this calculation produces a decimal value (e.g., 59/4 = 14.75), the ceiling of this number is taken (⌈1/4·59⌉ = 15). This is followed by a layer half the size of the target vector (
Gh2 = ⌈1/2·60⌉ = 30). The generator’s final hidden layer is the size of the vector plus one quarter (
Gh3 = ⌈1/4·60⌉ + 60 = 75). The discriminator, on the other hand, is composed of two hidden layers, with the first hidden layer being the same size as
Gh2, and the second hidden layer equivalent to
Gh1. Each hidden layer is followed by a batch normalization algorithm before being activated using the tanh function. Tanh works best considering the target domain is scaled to values between -1 and 1. Other components of the algorithm include a dropout layer (
p = 0.4) prior to the discriminator’s output and random normal kernel initializers in both models (discriminator σ = 0.1, generator σ = 0.7).
Experiments with loss functions and optimization algorithms showed a significant improvement in performance using
LSGAN and
WGAN variants when compared with vanilla GAN’s binary cross-entropy (rTOST
p < 0.05,
Table 3 with more details explained in
Section 3.2). All three GANs were able to generate realistic distributions, with
WGAN-GP outperforming
WGAN in some cases (
Table 3).
LSGAN additionally worked best when using Adam optimization (α = 0.0002, β
1 = 0.5), while
WGAN and
WGAN-GP excelled using RMSprop (α = 0.00005).
The optimal batch number was found at 16. This allowed the discriminator enough data to objectively evaluate performance and, thus, resulted in more efficient weight updates for the generator. The number of epochs, however, were highly dependent on the number of individuals used for training. Finally, the dimensionality of latent space was also found to be conditioned by the size of the target domain, as will be explained in continuation.
3.2. Experiments with Dimensionality and Sample Size
Initial trials with latent space found ℝ
50 to produce the best results on average, especially in the case of DS2 (results of which have already been reported in
Table 2). Nevertheless, interesting patterns emerged when experimenting with larger and smaller latent vector inputs. Starting with the case of the smallest target domain (DS1,
Table 4), a significant negative correlation was detected when observing rTOST
p values compared with the size of the latent vector over numerous runs (Kendall’s τ = −0.44,
p = 0.001). This is also true when considering rTOST absolute difference values (τ = −0.41,
p = 0.003). This correlation highlights larger ℝ
ns to work best when working with smaller target domains. When training on larger feature spaces (e.g., DS3,
Table 5), correlations proved insignificant for both rTOST
p values (Kendall’s τ = −0.21,
p = 0.13) and absolute difference calculations (τ = −0.29,
p = 0.15). Nevertheless, while correlations remain insignificant, smaller latent vectors were seen to create more predictable and stable data (
Figure 5).
In most experiments, 400 epochs were considered enough for GANs to produce realistic data. Moreover, the best results of each GAN began appearing after approximately 100-130 epochs. Performance significantly decreased, however, when trained on the same number of epochs using less data. To test this, subsets of each dataset were taken for experimentation (e.g., 30 out of 60 samples from DS2,
Table 6). On all accounts, significant correlations were detected, finding smaller datasets to need more training time in order to obtain optimal results (Pearson’s
r = 0.65,
p = 0.0005). Likewise,
LSGAN appeared to be the model least affected by dataset size, producing the most realistic distributions in each of the cases (
Table 6). While training GANs using 400 epochs is still able to produce realistic data on small datasets, when considering the optimal number of epochs, increasing this number to 1000 produces a significant improvement in results.
3.3. Data Augmentation Results
3.3.1. General GAN Performance
All three GANs are highly successful in replicating sample distributions, effectively augmenting each of the distributions without too much distortion (
Figure 7,
Tables S1–S6). While evaluating standalone synthetic data creates some confusion, seen in some deviations of synthetic central tendency values and IR intervals (
Tables S1, S3 and S5), the true value of GANs are observed when considering the augmented sample as a whole (
Tables S2, S4 and S6).
In most cases, it can be seen how even the worst performing algorithms are able to maintain the central tendency of samples while boosting the variance represented. It is also important to highlight that, while in some cases central tendency can be seen to deviate slightly from the original distribution (e.g.,
LSGAN on DS2,
Table S2), this is normally only true of one PC score and is still insignificant (rTOST
p < 0.05). Some algorithms are also seen to affect the normality of sample distributions, creating distortion that is reflected in increased sample skewness. Nevertheless, these distortions are still unable to modify the general magnitude of similarities between synthetic and real data.
The greatest value of GANs can, therefore, be seen in increases in overall sample variance without significant distortion of the real sample’s distribution. Deviation values and IR intervals increase, representing more variability without significantly shifting central tendency and without generating outliers. This shows how each algorithm is able to essentially “fill in the gaps” for each distribution while staying true to the original domain.
If GAN performance were to be compared with more traditional augmentation procedures, such as bootstrap, GANs can be seen to smooth out the distribution curve (
Figure 8), creating a more general and complete mapping of the target domain. Bootstrapping procedures, on the other hand, tend to exaggerate gaps in the distribution. This can mostly be characterized by noticeable modifications to sample kurtosis while maintaining the general variation (
Table 7).
3.3.2. Conditional GAN Performance
CGAN presented limited success when augmenting datasets, with only Wasserstein Gradient Penalty loss succeeding in overcoming mode collapse. Nevertheless, CGAN was still able to generate data with insignificant differences (
Table 8), successfully augmenting the targeted datasets (
Table 9 and
Figure 9).
When taking a closer look at the performance of CGAN, however, it is important to note that, while the magnitude of differences between synthetic and real data are insignificant, CGAN distorts the original distribution to a greater degree (
Figure 9). In both samples, CGAN deviates greatly from the target central tendency (
Table 9) and appears to shift the general skew of the distribution (
Figure 9). When using GANs to augment each of the samples separately, however, the generated data is arguably truer to the original domain. This is not to say, however, that CGANs are unable to augment feature spaces successfully. With the right configuration, CGANs are likely to reach similar results to traditional GANs. This, however, goes beyond the scope of the present study.
4. Discussion
Many algorithms require large amounts of data in order to efficiently extract information, a task which is particularly difficult when considering data derived from the fossil record. To confront this topic, this study presents a new integration of AIAs into archaeological and paleontological sciences. Here GANs have been shown to be a new and valuable tool for the modelling and augmentation of GM data. Moreover, these algorithms can additionally be employed on a number of different types of datasets and applications; whether this be the handling of paleoanthropological, biological, taphonomic or lithic specimens via GM landmark data.
To demonstrate this latter point, and applying typical geometric morphometric techniques for classification on the O’Higgins and Dryden [
75] Hominoid skull dataset, the present study is able to increment balanced accuracy of traditional LDA up to ca. 5% (
Figure 10a) with a significant increase in generalization (
Figure 10b,c). For this demonstration LDA was trained using a traditional approach [
11], as well as an augmented approach based on Machine Teaching [
43]. It can be seen how applying Machine Teaching using 100 realistic synthetic points per sample for training, and the original data was used for testing, helps the generalization process (
Figure 10b) while providing clearer boundaries for each of the sample domains (
Figure 10c).
It is important to point out, however, that this is not the solution to all sample-size related issues, and a number of components have to be discussed before more advanced applications can be carried out.
Missing data and the availability of fossil finds are a major handicap in prehistoric research. This is increasingly relevant when considering fossils of older ages, such as individuals of the
Australopithecus, Paranthropus and early
Homo genera. In a number of cases, for example, the representation of
Australopithecine or
Homo erectus/ergaster specimens may not even surpass 10 individuals [
76,
77,
78], while
Homo sapiens specimens are in abundance. In these cases, and in accordance with the data presented here, GANs would not be able to successfully augment the targeted minority distributions from scratch. Other options, however, could entail the use of algorithms for pre-processing, using variations of the Synthetic Minority Oversampling Technique (SMOTE & Borderline-SMOTE) [
79,
80,
81], or the adapted version Adaptive Synthetic Sampling (ADASYN) [
82].
Both SMOTE and ADASYN are useful, easy to implement algorithms that augment minority samples in imbalanced datasets. SMOTE generates synthetic data in the feature spaces that join data points (e.g., according to
k nearest-neighbor theory), thus filling in regions of the target domain [
79,
80,
81]. ADASYN takes this a step further by modelling on sample distributions based on data-density [
82]. Both are valuable algorithms that have become popular in imbalanced learning tasks, generally improving predictive model generalization. Nevertheless, their application should be confronted conservatively.
Preliminary experiments within this study found that resampling via bootstrapping prior to the training of GANs resulted in severe mode collapse. This can be theoretically explained by the manner in which bootstrapping is over-inflating the domain and highlighting very specific regions which the model then learns from. This results in overfitting as the model is repeatedly learning to map out the same value multiple times, boosting the probability of mode collapse through an enhanced lack of variation in the original trainset. Considering how SMOTE and ADASYN produce more “meaningful” data [
44,
79], these algorithms are more likely to aid the training process rather than produce the adverse effect. Nevertheless, overuse of SMOTE/ADASYN is likely to have a similar effect to bootstrapping, where linear regions of feature space between data points are enhanced while other regions are left empty.
Through this, the current study proposes that a conservative use of SMOTE or ADASYN variants prior to the training of GANs may be able to boost performance on overly-scarce datasets (e.g., the cases of [
76,
77,
78]). This practice would be able to augment minority samples to a suitable threshold (
n = 30), preparing the dataset for more complex generative modelling and enabling an improved generalization of any predictive models used in analyses that follow.
From a similar perspective, the use of Bayesian Inference Algorithms such as Markov Chain Monte Carlo (MCMC) and Metropolis-Hastings algorithms have also been known to effectively model from multiple types of probability distributions [
83,
84,
85]. In some cases, it may be possible to use these approaches to sample from the probability distribution at hand, and produce simulated information from the target distribution which would essentially be more realistic than simple bootstrap approaches. Further research into how these approaches may be applied could provide a powerful insight into GAN alternatives for different types of numerical data in GM.
In the general context of computational modelling, common criticism of neural network applications in archaeology and paleoanthropology argue that GM datasets are generally insufficient for the training of AIAs. This is based on the fact that most DL algorithms require much more data to avoid overfitting. From this point of view, why would training a GAN on such little data be any different? The present study proves that this is not an issue, considering how, with only 30 individuals, GANs are still able to produce highly realistic synthetic data (LSGAN rTOST p = 3.8 × 10−22).
In common DL literature, state-of-the-art models are reported to obtain ≈80% accuracy when trained on thousands to millions of specimens [
25]. It is important to consider, however, that in the majority of these cases AIAs are trained on
images (i.e., computer vision applications). To provide an example, Karras et al. [
54] present a GAN capable of producing hyper-realistic fake images of people’s faces, building from a subset of the CelebA-HQ dataset using ≈30,000 images. Two main components must be considered in order to understand why such a large dataset is required for their model;
Karras et al. [
54] present a GAN capable of producing high resolution 1024x1024 pixel RGB images. In computer vision applications, each image is conceptualized as a multidimensional numeric matrix (i.e., a tensor). Each of the numbers within the tensor can essentially be considered a variable, resulting in a dataset of approx. three million variables per individual photo.
In order to efficiently map out these three million numeric values, the featured GAN uses progressively growing convolutional layers (nº layers ≈ 60) with 23.1 million adjustable parameters.
The present study uses feature spaces that have already undergone dimensionality reduction derived from GM landmark data. In the case of the largest dataset, this results in a target vector of 60 variables that need to be generated. The present study additionally only uses fully connected layers with no convolutional filters, resulting in a model of <11,000 adjustable parameters. A GAN targeting three million values with 23.1 million parameters would thus require a far larger dataset than one targeting 60 values with 11 thousand parameters, explaining why with just 30 specimens, GAN convergence is still possible.
Regardless of the mathematics behind DL theory, the statistical results presented here provide enough empirical evidence to argue the value of the proposed GAN with as little as 30 individuals. Nevertheless, even in cases where datasets are too scarce for GANs to be developed from scratch, pre-trained models can be adjusted to different domains via multiple DL techniques. This arguably opens up new possibilities for the incorporation of Transfer Learning into GMs [
25].
Finally, it is important to highlight how no absolute protocol can be established for generative modelling of any type. DL practitioners are usually required to adapt their model according to the dataset at hand, using the best practices established in other studies as a baseline from which to work from. Under this premise, recommendations established for the augmentation of GM datasets using GANs can be listed as follows:
Best results are obtained when scaling the target domain to values between −1 and 1.
Hidden layer densities should be adjusted according to the number of dimensions within the target domain (
Figure 4). Tanh activation functions in both the generator and the discriminator are recommended.
Dropout, batch normalization and kernel initializers (discriminator σ = 0.1, generator σ = 0.7) are recommended to regulate the learning process and avoid mode collapse.
The Adam optimization algorithm is recommended when using Least-Square loss, while RMSProp is more efficient when using the Wasserstein (WGAN or WGAN-GP) function. A minimum batch size of 16 obtains the best results.
LSGAN is recommended when training data is limited, increasing the number of epochs to at least 1000.
WGAN and WGAN-GP work best on larger datasets, while approximately 400 epochs are usually enough to produce realistic data.
The smaller the target domain, the larger the latent vector required for generator input.
For conditional augmentation, optimal results are obtained by training GANs on each sample separately, rather than using CGANs.