A Novel Deep Learning Method Based on an Overlapping Time Window Strategy for Brain–Computer Interface-Based Stroke Rehabilitation
Abstract
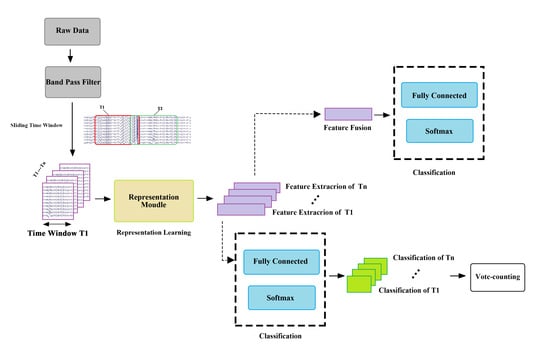
:1. Introduction
2. Materials and Methods
2.1. Experimental Protocol
2.2. Data Acquisition and Preprocessing
2.3. Overlapping Time Window
2.4. Graph Isomorphism Network Model
2.4.1. Graph Data Construction
2.4.2. Graph Isomorphism Network
2.5. CNN
2.6. LSTM
2.7. Evaluation Procedures
3. Results and Discussion
3.1. Overall Performance
3.2. Effects of Window Size
3.3. Generic Performance of BCI
3.4. The Visualization of Feature Distribution
3.5. The Impact of the Number of Network Layers
3.6. The Visualization of Accuracy on Time Window
3.7. Study of Cortical Activity on the Time Window
3.8. Limitations in Current Work
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Broussalis, E.; Killer, M.; McCoy, M.; Harrer, A.; Trinka, E.; Kraus, J. Current therapies in ischemic stroke. Part A. Recent developments in acute stroke treatment and in stroke prevention. Drug Discov. Today 2012, 17, 296–309. [Google Scholar] [CrossRef] [PubMed]
- Thieme, H.; Mehrholz, J.; Pohl, M.; Behrens, J.; Dohle, C. Mirror therapy for improving motor function after stroke. Stroke 2013, 44, e1–e2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saposnik, G.; Mcilroy, W.E.; Teasell, R.; Thorpe, K.E.; Bayley, M.; Cheung, D.; Mamdani, M.; Hall, J.; Cohen, L.G. Effectiveness of virtual reality using Wii gaming technology in stroke rehabilitation: A pilot randomized clinical trial and proof of principle. Stroke 2010, 41, 1477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rimmer, J.H.; Wang, E. Aerobic Exercise Training in Stroke Survivors. Top. Stroke Rehabil. 2005, 12, 17–30. [Google Scholar] [CrossRef]
- Ang, K.K.; Guan, C.; Phua, K.S.; Wang, C.; Zhou, L.; Tang, K.Y.; Ephraim Joseph, G.J.; Kuah, C.W.K.; Chua, K.S.G. Brain-computer interface-based robotic end effector system for wrist and hand rehabilitation: Results of a three-armed randomized controlled trial for chronic stroke. Front. Neuroeng. 2014, 7, 30. [Google Scholar] [CrossRef] [Green Version]
- Cervera, M.A.; Soekadar, S.R.; Ushiba, J.; Millán, J.D.R.; Liu, M.; Birbaumer, N.; Garipelli, G. Brain-computer interfaces for post-stroke motor rehabilitation: A meta-analysis. Ann. Clin. Transl. Neurol. 2018, 5, 651–663. [Google Scholar] [CrossRef]
- Mane, R.; Chouhan, T.; Guan, C. BCI for stroke rehabilitation: Motor and beyond. J. Neural. Eng. 2020, 17, 041001. [Google Scholar] [CrossRef]
- Pfurtscheller, G.; Neuper, C. Motor imagery activates primary sensorimotor area in humans. Neurosci. Lett. 1997, 239, 65–68. [Google Scholar] [CrossRef]
- Philip, G.R.; Daly, J.J.; Príncipe, J.C. Topographical measures of functional connectivity as biomarkers for post-stroke motor recovery. J. Neuroeng. Rehabil. 2017, 14, 67. [Google Scholar] [CrossRef]
- Xiao, X.; Xu, M.; Jin, J.; Wang, Y.; Jung, T.P.; Ming, D. Discriminative canonical pattern matching for single-trial classification of ERP components. IEEE Trans. Biomed. Eng. 2019, 67, 2266–2275. [Google Scholar] [CrossRef]
- Ramoser, H.; Muller-Gerking, J.; Pfurtscheller, G. Optimal spatial filtering of single trial EEG during imagined hand movement. IEEE Trans. Rehabil. Eng. 2000, 8, 441–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ang, K.K.; Chin, Z.Y.; Zhang, H.; Guan, C. Filter Bank Common Spatial Pattern (FBCSP) in Brain-Computer Interface. In Proceedings of the 2008 IEEE International Joint Conference on Neural Networks (IEEE World Congress on Computational Intelligence), Hong Kong, China, 1–8 June 2008; pp. 2390–2397. [Google Scholar]
- Geethanjali, P.; Mohan, Y.K.; Sen, J. Time Domain Feature Extraction and Classification of EEG Data for Brain Computer Interface. In Proceedings of the 2012 9th International Conference on Fuzzy Systems and Knowledge Discovery, Chongqing, China, 29–31 May 2012; pp. 1136–1139. [Google Scholar]
- Chen, S.; Shu, X.; Wang, H.; Ding, L.; Fu, J.; Jia, J. The differences between motor attempt and motor imagery in brain-computer interface accuracy and event-related desynchronization of patients with hemiplegia. Front. Neurorobot. 2021, 15, 706630. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.J.; Jia, T.; Li, C.; Li, T.; Qian, C.; Li, Z.; Pan, Y.; Ji, L. CNN-Based Prognosis of BCI Rehabilitation Using EEG From First Session BCI Training. IEEE Trans. Neural Syst. Rehabil. Eng. 2021, 29, 1936–1943. [Google Scholar] [CrossRef]
- Liang, F.Y.; Zhong, C.H.; Zhao, X.; Castro, D.L.; Chen, B.; Gao, F.; Liao, W.H. Online Adaptive and Lstm-Based Trajectory Generation of Lower Limb Exoskeletons for Stroke Rehabilitation. In Proceedings of the 2018 IEEE International Conference on Robotics and Biomimetics (ROBIO), Kuala Lumpur, Malaysia, 12–15 December 2018; pp. 27–32. [Google Scholar]
- Jin, J.; Sun, H.; Daly, I.; Li, S.; Liu, C.; Wang, X.; Cichocki, A. A Novel Classification Framework Using the Graph Representations of Electroencephalogram for Motor Imagery Based Brain-Computer Interface. IEEE Trans. Neural. Syst. Rehabil. Eng. 2021, 30, 20–29. [Google Scholar] [CrossRef]
- Wang, F.; Zhong, S.h.; Peng, J.; Jiang, J.; Liu, Y. Data Augmentation for Eeg-Based Emotion Recognition with Deep Convolutional Neural Networks. In Proceedings of the International Conference on Multimedia Modeling; Springer: Berlin/Heidelberg, Germany, 2018; pp. 82–93. [Google Scholar]
- A, I.U.; B, M.H.; B, E.U.H.Q.; B, H.A. An automated system for epilepsy detection using EEG brain signals based on deep learning approach. Expert Syst. Appl. 2018, 107, 61–71. [Google Scholar]
- Zhang, G.; Davoodnia, V.; Sepas-Moghaddam, A.; Zhang, Y.; Etemad, A. Classification of hand movements from EEG using a deep attention-based LSTM network. IEEE Sens. J. 2019, 20, 3113–3122. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, K.G.; Schirrmeister, R.T.; Ball, T. EEG-GAN: Generative adversarial networks for electroencephalograhic (EEG) brain signals. arXiv 2018, arXiv:1806.01875. [Google Scholar]
- Truong, N.D.; Zhou, L.; Kavehei, O. Semi-Supervised Seizure Prediction with Generative Adversarial Networks. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019. [Google Scholar]
- Zhou, J.; Cui, G.; Hu, S.; Zhang, Z.; Yang, C.; Liu, Z.; Wang, L.; Li, C.; Sun, M. Graph neural networks: A review of methods and applications. AI Open 2020, 1, 57–81. [Google Scholar] [CrossRef]
- Zhang, T.; Wang, X.; Xu, X.; Chen, C.P. GCB-Net: Graph convolutional broad network and its application in emotion recognition. IEEE Trans. Affect. Comput. 2019, 13, 379–388. [Google Scholar] [CrossRef]
- Shen, L.; Sun, M.; Li, Q.; Li, B.; Pan, Z.; Lei, J. Multiscale Temporal Self-Attention and Dynamical Graph Convolution Hybrid Network for EEG-Based Stereogram Recognition. IEEE Trans. Neural. Syst. Rehabil. Eng. 2022, 30, 1191–1202. [Google Scholar] [CrossRef]
- Song, T.; Zheng, W.; Song, P.; Cui, Z. EEG emotion recognition using dynamical graph convolutional neural networks. IEEE Trans. Affect. Comput. 2018, 11, 532–541. [Google Scholar] [CrossRef] [Green Version]
- Li, Y. A Survey of EEG Analysis based on Graph Neural Network. In Proceedings of the 2021 2nd International Conference on Electronics, Communications and Information Technology (CECIT), Sanya, China, 27–29 December 2021; pp. 151–155. [Google Scholar]
- Xu, K.; Hu, W.; Leskovec, J.; Jegelka, S. How powerful are graph neural networks? arXiv 2018, arXiv:1810.00826. [Google Scholar]
- Lun, X.; Yu, Z.; Chen, T.; Wang, F.; Hou, Y. A simplified CNN classification method for MI-EEG via the electrode pairs signals. Front. Hum. Neurosci. 2020, 14, 338. [Google Scholar] [CrossRef] [PubMed]
- Acharya, U.R.; Oh, S.L.; Hagiwara, Y.; Tan, J.H.; Adeli, H. Deep convolutional neural network for the automated detection and diagnosis of seizure using EEG signals. Comput. Biol. Med. 2018, 100, 270–278. [Google Scholar] [CrossRef]
- Pascanu, R.; Gulcehre, C.; Cho, K.; Bengio, Y. How to Construct Deep Recurrent Neural Networks. arXiv 2013, arXiv:1312.6026. [Google Scholar]
- Wolpaw, J.R.; Birbaumer, N.; Heetderks, W.J.; McFarland, D.J.; Peckham, P.H.; Schalk, G.; Donchin, E.; Quatrano, L.A.; Robinson, C.J.; Vaughan, T.M. Brain-computer interface technology: A review of the first international meeting. IEEE Trans. Rehabil. Eng. 2000, 8, 164–173. [Google Scholar] [CrossRef]
- Rasheed, S.; Mumtaz, W. Classification of Hand-Grasp Movements of Stroke Patients using EEG Data. In Proceedings of the 2021 International Conference on Artificial Intelligence (ICAI), Islamabad, Pakistan, 5–7 April 2021; pp. 86–90. [Google Scholar]
- Gers, F.A.; Schmidhuber, J.; Cummins, F. Learning to forget: Continual prediction with LSTM. Neural Comput. 2000, 12, 2451–2471. [Google Scholar] [CrossRef]
- Huang, X.; Xu, Y.; Hua, J.; Yi, W.; Yin, H.; Hu, R.; Wang, S. A Review on Signal Processing Approaches to Reduce Calibration Time in EEG-Based Brain–Computer Interface. Front. Neurosci. 2021, 15, 1066. [Google Scholar] [CrossRef]
- Khalaf, A.; Sejdic, E.; Akcakaya, M. Common spatial pattern and wavelet decomposition for motor imagery EEG-fTCD brain-computer interface. J. Neurosci. Methods 2019, 320, 98–106. [Google Scholar] [CrossRef]
- Zeng, X.; Zhu, G.; Yue, L.; Zhang, M.; Xie, S. A feasibility study of ssvep-based passive training on an ankle rehabilitation robot. J. Healthc. Eng. 2017, 2017, 6819056. [Google Scholar] [CrossRef] [Green Version]
- Majidov, I.; Whangbo, T. Efficient Classification of Motor Imagery Electroencephalography Signals Using Deep Learning Methods. Sensors 2019, 19, 1736. [Google Scholar] [CrossRef] [Green Version]
- Levine, B.; Schweizer, T.; O’Connor, C.; Turner, G.; Gillingham, S.; Stuss, D.; Manly, T.; Robertson, I. Rehabilitation of Executive Functioning in Patients with Frontal Lobe Brain Damage with Goal Management Training. Front. Hum. Neurosci. 2011, 5, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, S.; Zeng, Y.; Lei, W.; Wei, L.; Liu, Z.; Zhang, S.; Yang, J.; Wen, W. A Study of the Brain Abnormalities of Post-Stroke Depression in Frontal Lobe Lesion. Sci. Rep. 2017, 7, 13203. [Google Scholar]
- Xu, R.; Jiang, N.; Vuckovic, A.; Hasan, M.; Mrachacz-Kersting, N.; Allan, D.; Fraser, M.; Nasseroleslami, B.; Conway, B.; Dremstrup, K. Movement-related cortical potentials in paraplegic patients: Abnormal patterns and considerations for BCI-rehabilitation. Front. Neuroeng. 2014, 7, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, J.; Yin, E.; Jin, J.; Saab, R.; Daly, I.; Wang, X.; Hu, D.; Cichocki, A. Towards correlation-based time window selection method for motor imagery BCIs. Neural. Netw. 2018, 102, 87–95. [Google Scholar] [CrossRef] [PubMed]










| Subject | Sex | Age | Affected Limb | Stroke Stage |
|---|---|---|---|---|
| Sub1 | Male | 31 | Right | Subacute |
| Sub2 | Male | 40 | Left | Subacute |
| Sub3 | Male | 42 | Right | Subacute |
| Sub4 | Male | 47 | Right | Subacute |
| Sub5 | Male | 36 | Right | Subacute |
| Sub6 | Male | 30 | Right | Subacute |
| Sub7 | Male | 65 | Left | Subacute |
| Mean | - | 41.6 ± 12.0 | - | - |
| Subjects | Accuracy | |||||||
|---|---|---|---|---|---|---|---|---|
| CSP | FBCSP | GIN&VS | LSTM&VS | CNN&VS | GIN&FFS | LSTM&FFS | CNN&FFS | |
| sub1 | 0.798 ± 0.079 | 0.614 ± 0.096 | 0.914 ± 0.061 | 0.979 ± 0.025 | 0.944 ± 0.059 | 0.931 ± 0.049 | 0.980 ± 0.017 | 0.934 ± 0.058 |
| sub2 | 0.700 ± 0.064 | 0.578 ± 0.096 | 0.722 ± 0.069 | 0.772 ± 0.106 | 0.741 ± 0.065 | 0.746 ± 0.060 | 0.806 ± 0.111 | 0.757 ± 0.057 |
| sub3 | 0.771 ± 0.077 | 0.627 ± 0.094 | 0.902 ± 0.075 | 0.938 ± 0.044 | 0.932 ± 0.047 | 0.919 ± 0.073 | 0.949 ± 0.042 | 0.940 ± 0.046 |
| sub4 | 0.804 ± 0.072 | 0.635 ± 0.157 | 0.938 ± 0.038 | 0.993 ± 0.011 | 0.966 ± 0.024 | 0.952 ± 0.030 | 0.994 ± 0.010 | 0.970 ± 0.016 |
| sub5 | 0.724 ± 0.054 | 0.644 ± 0.057 | 0.711 ± 0.066 | 0.808 ± 0.071 | 0.762 ± 0.076 | 0.757 ± 0.073 | 0.841 ± 0.068 | 0.769 ± 0.085 |
| sub6 | 0.687 ± 0.040 | 0.630 ± 0.053 | 0.744 ± 0.055 | 0.801 ± 0.037 | 0.764 ± 0.058 | 0.777 ± 0.055 | 0.825 ± 0.029 | 0.755 ± 0.050 |
| sub7 | 0.707 ± 0.082 | 0.677 ± 0.090 | 0.798 ± 0.078 | 0.899 ± 0.066 | 0.845 ± 0.085 | 0.842 ± 0.081 | 0.909 ± 0.062 | 0.856 ± 0.084 |
| Mean | 0.742 | 0.629 | 0.819 | 0.884 | 0.851 | 0.846 | 0.901 | 0.854 |
| Group1 | 0.736 ± 0.056 | 0.629 ± 0.015 | 0.790 ± 0.108 | 0.865 ± 0.099 | 0.823 ± 0.105 | 0.822 ± 0.095 | 0.884 ± 0.088 | 0.820 ± 0.100 |
| Group2 | 0.746 ± 0.050 | 0.630 ± 0.041 | 0.840 ± 0.099 | 0.901 ± 0.094 | 0.871 ± 0.101 | 0.865 ± 0.092 | 0.915 ± 0.080 | 0.881 ± 0.096 |
| Method | Comparsion Method | T | df | Sig. (2-Tailed) |
|---|---|---|---|---|
| GIN&VS | GIN&FFS | −5.451 | 6 | 0.002 |
| CNN&FFS | −5.345 | 6 | 0.002 | |
| LSTM&FFS | −7.528 | 6 | <0.001 | |
| CNN&VS | −6.705 | 6 | 0.001 | |
| LSTM&VS | −7.140 | 6 | <0.001 | |
| CNN&VS | GIN&FFS | 1.146 | 6 | 0.295 |
| CNN&FFS | −0.999 | 6 | 0.356 | |
| LSTM&FFS | −5.976 | 6 | 0.001 | |
| LSTM&VS | −5.805 | 6 | 0.001 | |
| LSTM&VS | GIN&FFS | 6.823 | 6 | <0.001 |
| CNN&FFS | 4.333 | 6 | 0.005 | |
| LSTM&FFS | −3.527 | 6 | 0.012 | |
| GIN&FFS | CNN&FFS | −1.501 | 6 | 0.184 |
| LSTM&FFS | −8.477 | 6 | <0.001 | |
| CNN&FFS | LSTM&FFS | −5.400 | 6 | 0.002 |
| Window Length | Method | Accuracy | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Sub1 | Sub2 | Sub3 | Sub4 | Sub5 | Sub6 | Sub7 | Mean | ||
| 70 | GIN&VS | 0.929 ± 0.061 | 0.764 ± 0.069 | 0.915 ± 0.075 | 0.942 ± 0.038 | 0.760 ± 0.066 | 0.781 ± 0.055 | 0.858 ± 0.078 | 0.850 |
| GIN&FFS | 0.924 ± 0.049 | 0.755 ± 0.060 | 0.912 ± 0.073 | 0.947 ± 0.030 | 0.755 ± 0.073 | 0.782 ± 0.054 | 0.843 ± 0.081 | 0.845 | |
| CNN&VS | 0.94 ± 0.039 | 0.766 ± 0.076 | 0.931 ± 0.047 | 0.967 ± 0.021 | 0.777 ± 0.059 | 0.789 ± 0.054 | 0.871 ± 0.083 | 0.864 | |
| CNN&FFS | 0.939 ± 0.040 | 0.754 ± 0.079 | 0.929 ± 0.049 | 0.960 ± 0.018 | 0.761 ± 0.053 | 0.760 ± 0.059 | 0.856 ± 0.095 | 0.851 | |
| LSTM&VS | 0.985 ± 0.018 | 0.808 ± 0.101 | 0.951 ± 0.041 | 0.997 ± 0.008 | 0.844 ± 0.072 | 0.819 ± 0.036 | 0.917 ± 0.066 | 0.903 | |
| LSTM&FFS | 0.984 ± 0.014 | 0.801 ± 0.101 | 0.948 ± 0.038 | 0.995 ± 0.008 | 0.842 ± 0.078 | 0.812 ± 0.047 | 0.917 ± 0.078 | 0.900 | |
| 100 | GIN&VS | 0.925 ± 0.046 | 0.764 ± 0.058 | 0.916 ± 0.065 | 0.929 ± 0.039 | 0.746 ± 0.056 | 0.793 ± 0.053 | 0.844 ± 0.074 | 0.845 |
| GIN&FFS | 0.919 ± 0.053 | 0.755 ± 0.048 | 0.919 ± 0.069 | 0.935 ± 0.039 | 0.745 ± 0.070 | 0.784 ± 0.056 | 0.846 ± 0.080 | 0.843 | |
| CNN&VS | 0.938 ± 0.058 | 0.759 ± 0.057 | 0.923 ± 0.047 | 0.941 ± 0.030 | 0.755 ± 0.052 | 0.762 ± 0.053 | 0.852 ± 0.075 | 0.847 | |
| CNN&FFS | 0.925 ± 0.056 | 0.744 ± 0.064 | 0.920 ± 0.055 | 0.946 ± 0.029 | 0.748 ± 0.053 | 0.734 ± 0.057 | 0.830 ± 0.096 | 0.835 | |
| LSTM&VS | 0.982 ± 0.020 | 0.799 ± 0.095 | 0.943 ± 0.039 | 0.995 ± 0.008 | 0.807 ± 0.077 | 0.893 ± 0.038 | 0.816 ± 0.078 | 0.891 | |
| LSTM&FFS | 0.986 ± 0.017 | 0.792 ± 0.098 | 0.942 ± 0.040 | 0.995 ± 0.010 | 0.814 ± 0.084 | 0.898 ± 0.046 | 0.815 ± 0.089 | 0.892 | |
| 1000 | GIN | 0.810 ± 0.057 | 0.732 ± 0.062 | 0.814 ± 0.091 | 0.797 ± 0.051 | 0.696 ± 0.067 | 0.700 ± 0.037 | 0.731 ± 0.057 | 0.754 |
| CNN | 0.689 ± 0.084 | 0.625 ± 0.058 | 0.763 ± 0.013 | 0.643 ± 0.066 | 0.634 ± 0.051 | 0.612 ± 0.062 | 0.638 ± 0.067 | 0.658 | |
| LSTM | 0.674 ± 0.061 | 0.630 ± 0.051 | 0.719 ± 0.111 | 0.633 ± 0.075 | 0.644 ± 0.042 | 0.629 ± 0.042 | 0.646 ± 0.064 | 0.654 | |
| Conv Layers | Accuracy | |||||
|---|---|---|---|---|---|---|
| GIN&VS | CNN&VS | LSTM&VS | GIN&FFS | CNN&FFS | LSTM&FFS | |
| 1 layer | 0.803 ± 0.082 | 0.821 ± 0.091 | 0.882 ± 0.079 | 0.834 ± 0.074 | 0.791 ± 0.072 | 0.900 ± 0.065 |
| 2 layers | 0.819 ± 0.085 | 0.844 ± 0.086 | 0.884 ± 0.078 | 0.846 ± 0.075 | 0.846 ± 0.068 | 0.901 ± 0.066 |
| 3 layers | 0.816 ± 0.078 | 0.851 ± 0.083 | 0.883 ± 0.068 | 0.843 ± 0.082 | 0.854 ± 0.080 | 0.896 ± 0.069 |
| 4 layers | 0.806 ± 0.089 | 0.844 ± 0.088 | 0.882 ± 0.079 | 0.821 ± 0.078 | 0.847 ± 0.069 | 0.900 ± 0.072 |
| Mean | 0.810 | 0.840 | 0.882 | 0.836 | 0.835 | 0.900 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, L.; Wu, H.; Chen, S.; Dong, Y.; Zhu, C.; Jia, J.; Fan, C. A Novel Deep Learning Method Based on an Overlapping Time Window Strategy for Brain–Computer Interface-Based Stroke Rehabilitation. Brain Sci. 2022, 12, 1502. https://0-doi-org.brum.beds.ac.uk/10.3390/brainsci12111502
Cao L, Wu H, Chen S, Dong Y, Zhu C, Jia J, Fan C. A Novel Deep Learning Method Based on an Overlapping Time Window Strategy for Brain–Computer Interface-Based Stroke Rehabilitation. Brain Sciences. 2022; 12(11):1502. https://0-doi-org.brum.beds.ac.uk/10.3390/brainsci12111502
Chicago/Turabian StyleCao, Lei, Hailiang Wu, Shugeng Chen, Yilin Dong, Changming Zhu, Jie Jia, and Chunjiang Fan. 2022. "A Novel Deep Learning Method Based on an Overlapping Time Window Strategy for Brain–Computer Interface-Based Stroke Rehabilitation" Brain Sciences 12, no. 11: 1502. https://0-doi-org.brum.beds.ac.uk/10.3390/brainsci12111502