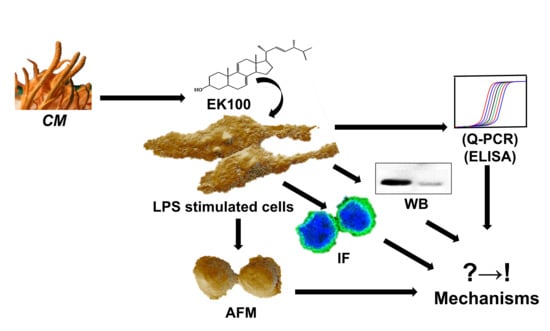
Ergosta-7,9(11),22-trien-3β-ol Attenuates Inflammatory Responses via Inhibiting MAPK/AP-1 Induced IL-6/JAK/STAT Pathways and Activating Nrf2/HO-1 Signaling in LPS-Stimulated Macrophage-like Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell Culture
2.3. Atomic Force Microscopy (AFM) Assay
2.4. Enzyme-Linked Immunosorbent Assay (ELISA)
2.5. Quantitative Polymerase Chain Reaction Assay (qPCR)
2.6. Western Blotting Analysis (WB)
2.7. Immunofluorescence Assay (IF)
2.8. Nrf2 siRNA Transfection Assay
2.9. Statistical Analysis
3. Results
3.1. EK100 Inhibited IL-6 and TNF-α Released in LPS-Stimulated RAW 264.7 Cells
3.2. EK100 Prevented the Morphological Change in LPS-Stimulated RAW 264.7 Cells
3.3. EK100 Suppressed the MAPK/AP-1 Pathways in LPS-Stimulated RAW 264.7 Cells
3.4. EK100 Inhibited the JAKs/STATs Pathways in LPS-Stimulated RAW 264.7 Cells
3.5. EK100 Activated the Nrf2/HO-1 Signaling Pathway in LPS-Stimulated Macrophage-like Cells
3.6. Nrf2 siRNA Reversed EK100 Activated the Nrf2/HO-1 Pathway in LPS-Stimulated Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AP-1 | Activator protein-1 |
| AFM | Atomic Force Microscopy |
| ARE | Antioxidant responsive element |
| Akt | Protein kinase B |
| BSA | Bovine serum albumin |
| CAT | Catalase |
| CLI-095 | Resatorvid; TAK-242 |
| CM | Cordyceps militaris |
| COX-2 | Cyclooxygenase-2 |
| Dexa | Dexamethasone |
| DAPI | 4′,6-diamidino-2-phenylindole |
| DMSO | Dimethyl sulfoxide |
| DMEM | Dulbecco’s Modified Eagle Medium |
| ECL | Enhanced chemiluminescence |
| ELISA | Enzyme-linked immunoassay |
| ERK | Extracellular-signal-regulated kinase |
| EK100 | Ergosta-7,9(11),22-trien-3β-ol |
| FBS | Fetal bovine serum |
| GPx | Glutathione peroxidase |
| HO-1 | Heme oxygenase-1 |
| IF | Immunofluorescence |
| IL | Interleukin |
| iNOS | Inducible nitric oxide synthase |
| IκB | Inhibitor kappa B |
| IKK | IκB kinase |
| JAK | Janus kinase |
| JNK | c-Jun N-terminal kinase |
| Keap-1 | Kelch-like ECH-associated protein |
| LPS | Lipopolysaccharide |
| MAPK | Mitogen-activated protein kinase |
| MD-2 | Myeloid differentiation-2 |
| MMP | Matrix metalloproteinase |
| MTT | 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide |
| NF-κB | Nuclear factor-κB |
| NO | Nitric oxide |
| NQO1 | NAD (P) H quinone oxidoreductase 1 |
| Nrf2 siRNA | Nrf2 Small interfering RNA |
| Nrf2 | Nuclear transcription factor erythroid 2-related factor 2 |
| p38 | p38 MAPK |
| p50 | NF-κB p50 |
| p65 | NF-κB p65 |
| PI3K | Phosphatidylinositol-3-kinase |
| PMA | Phorbol 12-myristate 13-acetate |
| PVDF | Polyvinylidene fluoride |
| Q-PCR | Quantitative real-time polymerase chain reaction |
| SDS-PAGE | Sodium dodecyl sulfate-polyacrylamide gel |
| SOD | Super oxidative dismutases |
| STAT | Signal transducer and activator of transcription |
| TLR4 | Toll-like receptor 4 |
| TNF-α | Tumor necrosis factor-α |
References
- Furman, D.; Campisi, J.; Verdin, E.; Carrera-Bastos, P.; Targ, S.; Franceschi, C.; Ferrucci, L.; Gilroy, D.W.; Fasano, A.; Miller, G.W.; et al. Chronic inflammation in the etiology of disease across the life span. Nat. Med. 2019, 25, 1822–1832. [Google Scholar] [CrossRef]
- Ahmed, S.M.U.; Luo, L.; Namani, A.; Wang, X.J.; Tang, X. Nrf2 signaling pathway: Pivotal roles in inflammation. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2017, 1863, 585–597. [Google Scholar] [CrossRef]
- Kobayashi, E.H.; Suzuki, T.; Funayama, R.; Nagashima, T.; Hayashi, M.; Sekine, H.; Tanaka, N.; Moriguchi, T.; Motohashi, H.; Nakayama, K.; et al. Nrf2 suppresses macrophage inflammatory response by blocking pro-inflammatory cytokine transcription. Nat. Commun. 2016, 7, 11624. [Google Scholar] [CrossRef]
- Mazur-Bialy, A.I.; Pocheć, E. The Time-Course of Antioxidant Irisin Activity: Role of the Nrf2/HO-1/HMGB1 Axis. Antioxidants 2021, 10, 88. [Google Scholar] [CrossRef]
- Leonard, M.; Ryan, M.P.; Watson, A.J.; Schramek, H.; Healy, E. Role of MAP kinase pathways in mediating IL-6 production in human primary mesangial and proximal tubular cells. Kidney Int. 1999, 56, 1366–1377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.W.; Sun, Y.M. The IL-6/JAK/STAT3 pathway: Potential therapeutic strategies in treating colorectal cancer (Review). Int. J. Oncol. 2014, 44, 1032–1040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levy, D.E.; Darnell, J.E., Jr. Stats: Transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 2002, 3, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Montoya, T.; Sánchez-Hidalgo, M.; Castejón, M.L.; Rosillo, M.; González-Benjumea, A.; Alarcón-de-la-Lastra, C. Dietary Oleocanthal Supplementation Prevents Inflammation and Oxidative Stress in Collagen-Induced Arthritis in Mice. Antioxidants 2021, 10, 650. [Google Scholar] [CrossRef]
- Muller, T.; Hengstermann, A. Nrf2: Friend and foe in preventing cigarette smoking-dependent lung disease. Chem. Res. Toxicol. 2012, 25, 1805–1824. [Google Scholar] [CrossRef] [PubMed]
- Das, S.K.; Masuda, M.; Sakurai, A.; Sakakibara, M. Medicinal uses of the mushroom Cordyceps militaris: Current state and prospects. Fitoterapia 2010, 81, 961–968. [Google Scholar] [CrossRef]
- Hsieh, W.-T.; Hsu, M.-H.; Lin, W.-J.; Xiao, Y.-C.; Lyu, P.-C.; Liu, Y.-C.; Lin, W.-Y.; Kuo, Y.-H.; Chung, J.-G. Ergosta-7, 9 (11), 22-trien-3β-ol Interferes with LPS Docking to LBP, CD14, and TLR4/MD-2 Co-Receptors to Attenuate the NF-κB Inflammatory Pathway In Vitro and Drosophila. Int. J. Mol. Sci. 2021, 22, 6511. [Google Scholar] [CrossRef]
- Wang, Y.H.; Chern, C.M.; Liou, K.T.; Kuo, Y.H.; Shen, Y.C. Ergostatrien-7,9(11),22-trien-3β-ol from Antrodia camphorata ameliorates ischemic stroke brain injury via downregulation of p65NF-κ-B and caspase 3, and activation of Akt/GSK3/catenin-associated neurogenesis. Food Funct. 2019, 10, 4725–4738. [Google Scholar] [CrossRef] [PubMed]
- Kuo, Y.H.; Lin, T.Y.; You, Y.J.; Wen, K.C.; Sung, P.J.; Chiang, H.M. Antiinflammatory and Antiphotodamaging Effects of Ergostatrien-3β-ol, Isolated from Antrodia camphorata, on Hairless Mouse Skin. Molecules 2016, 21, 1213. [Google Scholar] [CrossRef] [Green Version]
- Kao, S.-T.; Kuo, Y.-H.; Wang, S.-D.; Hong, H.-J.; Lin, L.-J. Analogous corticosteroids, 9A and EK100, derived from solid-state-cultured mycelium of Antrodia camphorata inhibit pro-inflammatory cytokine expression in macrophages. Cytokine 2018, 108, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.J.; Huang, S.S.; Lin, S.S.; Shao, Y.Y.; Chen, C.C.; Hou, W.C.; Kuo, Y.H. Analgesic effects and the mechanisms of anti-inflammation of ergostatrien-3beta-ol from Antrodia camphorata submerged whole broth in mice. J. Agric. Food Chem. 2010, 58, 7445–7452. [Google Scholar] [CrossRef] [PubMed]
- Pi, J.; Cai, H.; Yang, F.; Jin, H.; Liu, J.; Yang, P.; Cai, J. Atomic force microscopy based investigations of anti-inflammatory effects in lipopolysaccharide-stimulated macrophages. Anal. Bioanal. Chem. 2016, 408, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Jang, B.K.; Lee, J.W.; Choi, H.; Yim, S.V. Aronia melanocarpa Fruit Bioactive Fraction Attenuates LPS-Induced Inflammatory Response in Human Bronchial Epithelial Cells. Antioxidants 2020, 9, 816. [Google Scholar] [CrossRef]
- Luo, J.F.; Shen, X.Y.; Lio, C.K.; Dai, Y.; Cheng, C.S.; Liu, J.X.; Yao, Y.D.; Yu, Y.; Xie, Y.; Luo, P.; et al. Activation of Nrf2/HO-1 Pathway by Nardochinoid C Inhibits Inflammation and Oxidative Stress in Lipopolysaccharide-Stimulated Macrophages. Front. Pharmacol. 2018, 9, 911. [Google Scholar] [CrossRef]
- Hsieh, W.T.; Lin, H.Y.; Chen, J.H.; Lin, W.C.; Kuo, Y.H.; Wood, W.G.; Lu, H.F.; Chung, J.G. Latex of Euphorbia antiquorum-induced S-phase arrest via active ATM kinase and MAPK pathways in human cervical cancer HeLa cells. Environ. Toxicol. 2015, 30, 1205–1215. [Google Scholar] [CrossRef]
- Lin, Y.H.; Hsiao, Y.H.; Ng, K.L.; Kuo, Y.H.; Lim, Y.P.; Hsieh, W.T. Physalin A attenuates inflammation through down-regulating c-Jun NH2 kinase phosphorylation/Activator Protein 1 activation and up-regulating the antioxidant activity. Toxicol. Appl. Pharmacol. 2020, 402, 115115. [Google Scholar] [CrossRef]
- Jain, N.; Zhang, T.; Fong, S.L.; Lim, C.P.; Cao, X. Repression of Stat3 activity by activation of mitogen-activated protein kinase (MAPK). Oncogene 1998, 17, 3157–3167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neuder, L.E.; Keener, J.M.; Eckert, R.E.; Trujillo, J.C.; Jones, S.L. Role of p38 MAPK in LPS induced pro-inflammatory cytokine and chemokine gene expression in equine leukocytes. Vet. Immunol. Immunopathol. 2009, 129, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Nyati, K.K.; Masuda, K.; Zaman, M.M.; Dubey, P.K.; Millrine, D.; Chalise, J.P.; Higa, M.; Li, S.; Standley, D.M.; Saito, K.; et al. TLR4-induced NF-κB and MAPK signaling regulate the IL-6 mRNA stabilizing protein Arid5a. Nucleic Acids Res. 2017, 45, 2687–2703. [Google Scholar] [CrossRef] [PubMed]
- Pi, J.; Wang, Y.; Zhu, H.; Jin, H.; Jiang, J.; Yang, F.; Ma, C.W.; Hu, M.; Ma, F.; Cai, H.; et al. Immunomodulatory effects of polysaccharide compounds in macrophages revealed by high resolution AFM. Scanning 2016, 38, 792–801. [Google Scholar] [CrossRef] [Green Version]
- Sabio, G.; Davis, R.J. TNF and MAP kinase signalling pathways. Semin. Immunol. 2014, 26, 237–245. [Google Scholar] [CrossRef] [Green Version]
- Akira, S.; Takeda, K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004, 4, 499–511. [Google Scholar] [CrossRef]
- Kaminska, B. MAPK signalling pathways as molecular targets for anti-inflammatory therapy--from molecular mechanisms to therapeutic benefits. Biochim. Biophys. Acta 2005, 1754, 253–262. [Google Scholar] [CrossRef]
- Koyama, Y.; Kaidzu, S.; Kim, Y.C.; Matsuoka, Y.; Ishihara, T.; Ohira, A.; Tanito, M. Suppression of Light-Induced Retinal Degeneration by Quercetin via the AP-1 Pathway in Rats. Antioxidants 2019, 8, 79. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.C.; Chen, B.K.; Chang, W.C. Activating protein 1-mediated cyclooxygenase-2 expression is independent of N-terminal phosphorylation of c-Jun. Mol. Pharmacol. 2005, 67, 2057–2069. [Google Scholar] [CrossRef] [PubMed]
- Venugopal, R.; Jaiswal, A.K. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc. Natl. Acad. Sci. USA 1996, 93, 14960–14965. [Google Scholar] [CrossRef] [Green Version]
- He, F.; Ru, X.; Wen, T. NRF2, a Transcription Factor for Stress Response and Beyond. Int. J. Mol. Sci. 2020, 21, 4777. [Google Scholar] [CrossRef]
- Basu, A.; Das, A.S.; Sharma, M.; Pathak, M.P.; Chattopadhyay, P.; Biswas, K.; Mukhopadhyay, R. STAT3 and NF-κB are common targets for kaempferol-mediated attenuation of COX-2 expression in IL-6-induced macrophages and carrageenan-induced mouse paw edema. Biochem. Biophys. Rep. 2017, 12, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Hodge, D.R.; Hurt, E.M.; Farrar, W.L. The role of IL-6 and STAT3 in inflammation and cancer. Eur. J. Cancer 2005, 41, 2502–2512. [Google Scholar] [CrossRef] [PubMed]
- Zauberman, A.; Zipori, D.; Krupsky, M.; Ben-Levy, R. Stress activated protein kinase p38 is involved in IL-6 induced transcriptional activation of STAT3. Oncogene 1999, 18, 3886–3893. [Google Scholar] [CrossRef] [Green Version]
- Loboda, A.; Damulewicz, M.; Pyza, E.; Jozkowicz, A.; Dulak, J. Role of Nrf2/HO-1 system in development, oxidative stress response and diseases: An evolutionarily conserved mechanism. Cell. Mol. Life Sci. 2016, 73, 3221–3247. [Google Scholar] [CrossRef] [Green Version]
- Tu, W.; Wang, H.; Li, S.; Liu, Q.; Sha, H. The Anti-Inflammatory and Antioxidant Mechanisms of the Keap1/Nrf2/ARE Signaling Pathway in Chronic Diseases. Aging Dis. 2019, 10, 637–651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ooi, B.K.; Goh, B.H.; Yap, W.H. Oxidative Stress in Cardiovascular Diseases: Involvement of Nrf2 Antioxidant Redox Signaling in Macrophage Foam Cells Formation. Int. J. Mol. Sci. 2017, 18, 2336. [Google Scholar] [CrossRef] [Green Version]
- Bloom, D.A.; Jaiswal, A.K. Phosphorylation of Nrf2 at Ser40 by protein kinase C in response to antioxidants leads to the release of Nrf2 from INrf2, but is not required for Nrf2 stabilization/accumulation in the nucleus and transcriptional activation of antioxidant response element-mediated NAD(P)H:quinone oxidoreductase-1 gene expression. J. Biol. Chem. 2003, 278, 44675–44682. [Google Scholar] [CrossRef] [Green Version]
- Arunachalam, S.; Nagoor Meeran, M.F.; Azimullah, S.; Sharma, C.; Goyal, S.N.; Ojha, S. Nerolidol Attenuates Oxidative Stress, Inflammation, and Apoptosis by Modulating Nrf2/MAPK Signaling Pathways in Doxorubicin-Induced Acute Cardiotoxicity in Rats. Antioxidants 2021, 10, 984. [Google Scholar] [CrossRef] [PubMed]
- Jeyapaul, J.; Jaiswal, A.K. Nrf2 and c-Jun regulation of antioxidant response element (ARE)-mediated expression and induction of γ-glutamylcysteine synthetase heavy subunit gene. Biochem. Pharmacol. 2000, 59, 1433–1439. [Google Scholar] [CrossRef]
- Zhu, H.; Itoh, K.; Yamamoto, M.; Zweier, J.L.; Li, Y. Role of Nrf2 signaling in regulation of antioxidants and phase 2 enzymes in cardiac fibroblasts: Protection against reactive oxygen and nitrogen species-induced cell injury. FEBS Lett. 2005, 579, 3029–3036. [Google Scholar] [CrossRef] [Green Version]
- Lin, W.-C.; Deng, J.-S.; Huang, S.-S.; Lin, W.-R.; Wu, S.-H.; Lin, H.-Y.; Huang, G.-J. Anti-inflammatory activity of Sanghuangporus sanghuang by suppressing the TLR4-mediated PI3K/AKT/mTOR/IKKβ signaling pathway. RSC Adv. 2017, 7, 21234–21251. [Google Scholar] [CrossRef] [Green Version]
- Li, P.C.; Tu, M.J.; Ho, P.Y.; Jilek, J.L.; Duan, Z.; Zhang, Q.Y.; Yu, A.X.; Yu, A.M. Bioengineered NRF2-siRNA Is Effective to Interfere with NRF2 Pathways and Improve Chemosensitivity of Human Cancer Cells. Drug Metab. Dispos. 2018, 46, 2–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Sun, Y. Nrf2 is required for suppressing osteoclast RANKL-induced differentiation in RAW 264.7 cells via inactivating cannabinoid receptor type 2 with AM630. Regen. Ther. 2020, 14, 191–195. [Google Scholar] [CrossRef] [PubMed]












Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.-P.; Chen, D.-R.; Lin, W.-J.; Lin, Y.-H.; Chen, J.-Y.; Kuo, Y.-H.; Chung, J.-G.; Hsia, T.-C.; Hsieh, W.-T. Ergosta-7,9(11),22-trien-3β-ol Attenuates Inflammatory Responses via Inhibiting MAPK/AP-1 Induced IL-6/JAK/STAT Pathways and Activating Nrf2/HO-1 Signaling in LPS-Stimulated Macrophage-like Cells. Antioxidants 2021, 10, 1430. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox10091430
Huang Y-P, Chen D-R, Lin W-J, Lin Y-H, Chen J-Y, Kuo Y-H, Chung J-G, Hsia T-C, Hsieh W-T. Ergosta-7,9(11),22-trien-3β-ol Attenuates Inflammatory Responses via Inhibiting MAPK/AP-1 Induced IL-6/JAK/STAT Pathways and Activating Nrf2/HO-1 Signaling in LPS-Stimulated Macrophage-like Cells. Antioxidants. 2021; 10(9):1430. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox10091430
Chicago/Turabian StyleHuang, Yi-Ping, Dar-Ren Chen, Wen-Jen Lin, Yu-Hsien Lin, Jiann-Yeu Chen, Yueh-Hsiung Kuo, Jing-Gung Chung, Te-Chun Hsia, and Wen-Tsong Hsieh. 2021. "Ergosta-7,9(11),22-trien-3β-ol Attenuates Inflammatory Responses via Inhibiting MAPK/AP-1 Induced IL-6/JAK/STAT Pathways and Activating Nrf2/HO-1 Signaling in LPS-Stimulated Macrophage-like Cells" Antioxidants 10, no. 9: 1430. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox10091430