Optimization of Ultrasonic Extraction Parameters for the Recovery of Phenolic Compounds in Brown Seaweed: Comparison with Conventional Techniques
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Materials
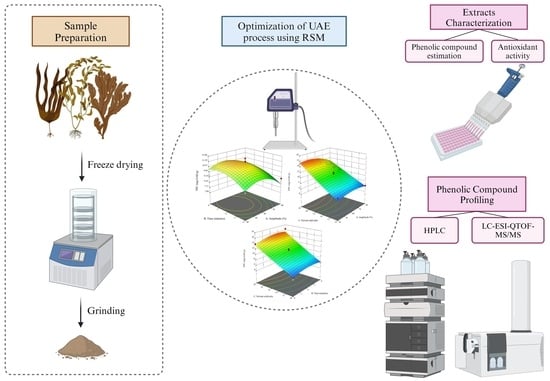
2.2. Seaweed Sample Preparation
2.3. Conventional Extraction of Free and Bound Phenolics
2.4. Ultrasonic Extraction of Free and Bound Phenolics
2.5. Experimental Design
2.6. Characterization of Free and Bound Phenolics from Conventional and Ultrasonic Extraction
2.6.1. Determination of Total Phenolic Content (TPC)
2.6.2. Determination of Total Flavonoid Content (TFC)
2.6.3. Determination of Total Condensed Tannin (TCT)
2.6.4. Analysis of Total Phlorotannin Content (TPhC)
2.6.5. DPPH Radical Scavenging Activity
2.6.6. ABTS Radical Scavenging Activity
2.6.7. Ferric-Reducing Antioxidant Power (FRAP)
2.6.8. Phosphomolybdate-Reducing Antioxidant Capacity (PRAC)
2.6.9. Ferrous Ion Chelating Activity (FICA)
2.7. Quantification of Phenolic Compounds by HPLC-PDA
2.8. Characterization of Phenolic Compounds by LC-ESI-QTOF-MS/MS Analysis
2.9. Statistical Analysis
3. Results and Discussion
3.1. Levels of Independent Extraction Variables for Free and Bound Phenolics Extraction
3.2. Effect of Experimental Model on Free and Bound Phenolics Extraction
3.3. Effect of Operational Parameters on the Extraction of Free and Bound Phenolics
3.4. Optimization and Verification of Extraction Conditions for Free and Bound Phenolics
3.5. Comparison between Conventional and Ultrasonic Extraction of Phenolic Compounds
3.6. Correlation among Phenolic Content and Antioxidant Activity
3.7. Heatmap Analysis of Phenolic Compounds
3.8. LC-ESI-QTOF-MS/MS Characterization
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cian, R.E.; Drago, S.R.; De Medina, F.S.; Martínez-Augustin, O. Proteins and Carbohydrates from Red Seaweeds: Evidence for Beneficial Effects on Gut Function and Microbiota. Mar. Drugs 2015, 13, 5358–5383. [Google Scholar] [CrossRef]
- Murai, U.; Yamagishi, K.; Kishida, R.; Iso, H. Impact of seaweed intake on health. Eur. J. Clin. Nutr. 2021, 75, 877–889. [Google Scholar] [CrossRef]
- Chichibu, H.; Yamagishi, K.; Kishida, R.; Maruyama, K.; Hayama-Terada, M.; Shimizu, Y.; Muraki, I.; Umesawa, M.; Cui, R.; Imano, H.; et al. Seaweed Intake and Risk of Cardiovascular Disease: The Circulatory Risk in Communities Study (CIRCS). J. Atheroscler. Thromb. 2021, 28, 1298–1306. [Google Scholar] [CrossRef]
- Brown, E.M.; Allsopp, P.J.; Magee, P.J.; Gill, C.I.; Nitecki, S.; Strain, C.R.; McSorley, E.M. Seaweed and human health. Nutr. Rev. 2014, 72, 205–216. [Google Scholar] [CrossRef]
- Cherry, P.; O’Hara, C.; Magee, P.J.; McSorley, E.M.; Allsopp, P.J. Risks and benefits of consuming edible seaweeds. Nutr. Rev. 2019, 77, 307–329. [Google Scholar] [CrossRef]
- Xie, C.; Lee, Z.J.; Ye, S.; Barrow, C.J.; Dunshea, F.R.; Suleria, H.A.R. A Review on Seaweeds and Seaweed-Derived Polysaccharides: Nutrition, Chemistry, Bioactivities, and Applications. Food Rev. Int. 2023, 1–36. [Google Scholar] [CrossRef]
- Visch, W.; Layton, C.; Hurd, C.L.; Macleod, C.; Wright, J.T. A strategic review and research roadmap for offshore seaweed aquaculture—A case study from southern Australia. Rev. Aquac. 2023, 15, 1467–1479. [Google Scholar] [CrossRef]
- Birkemeyer, C.; Lemesheva, V.; Billig, S.; Tarakhovskaya, E. Composition of Intracellular and Cell Wall-Bound Phlorotannin Fractions in Fucoid Algae Indicates Specific Functions of These Metabolites Dependent on the Chemical Structure. Metabolites 2020, 10, 369. [Google Scholar] [CrossRef]
- Phang, S.J.; Teh, H.X.; Looi, M.L.; Arumugam, B.; Fauzi, M.B.; Kuppusamy, U.R. Phlorotannins from brown algae: A review on their antioxidant mechanisms and applications in oxidative stress-mediated diseases. J. Appl. Phycol. 2023, 35, 867–892. [Google Scholar] [CrossRef]
- Besednova, N.N.; Andryukov, B.G.; Zaporozhets, T.S.; Kryzhanovsky, S.P.; Kuznetsova, T.A.; Fedyanina, L.N.; Makarenkova, I.D.; Zvyagintseva, T.N. Algae Polyphenolic Compounds and Modern Antibacterial Strategies: Current Achievements and Immediate Prospects. Biomedicines 2020, 8, 342. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Lopez, C.; Pereira, A.G.; Lourenço-Lopes, C.; Garcia-Oliveira, P.; Cassani, L.; Fraga-Corral, M.; Prieto, M.A.; Simal-Gandara, J. Main bioactive phenolic compounds in marine algae and their mechanisms of action supporting potential health benefits. Food Chem. 2021, 341, 128262. [Google Scholar] [CrossRef]
- Topuz, O.K.; Gokoglu, N.; Yerlikaya, P.; Ucak, I.; Gumus, B. Optimization of antioxidant activity and phenolic compound extraction conditions from red seaweed (Laurencia obtuse). J. Aquat. Food Prod. Technol. 2016, 25, 414–422. [Google Scholar] [CrossRef]
- Matos, G.S.; Pereira, S.G.; Genisheva, Z.A.; Gomes, A.M.; Teixeira, J.A.; Rocha, C.M.R. Advances in Extraction Methods to Recover Added-Value Compounds from Seaweeds: Sustainability and Functionality. Foods 2021, 10, 516. [Google Scholar] [CrossRef]
- Lee, Z.J.; Xie, C.; Ng, K.; Suleria, H.A.R. Unraveling the bioactive interplay: Seaweed polysaccharide, polyphenol and their gut modulation effect. Crit. Rev. Food Sci. Nutr. 2023, 1–24. [Google Scholar] [CrossRef]
- Subbiah, V.; Ebrahimi, F.; Agar, O.T.; Dunshea, F.R.; Barrow, C.J.; Suleria, H.A. Comparative Study on the Effect of Phenolics and Their Antioxidant Potential of Freeze-Dried Australian Beach-Cast Seaweed Species upon Different Extraction Methodologies. Pharmaceuticals 2023, 16, 773. [Google Scholar] [CrossRef]
- Wu, Y.; Gao, H.; Wang, Y.; Peng, Z.; Guo, Z.; Ma, Y.; Zhang, R.; Zhang, M.; Wu, Q.; Xiao, J. Effects of different extraction methods on contents, profiles, and antioxidant abilities of free and bound phenolics of Sargassum polycystum from the South China Sea. J. Food Sci. 2022, 87, 968–981. [Google Scholar] [CrossRef]
- Duan, X.; Subbiah, V.; Xie, C.; Agar, O.T.; Barrow, C.J.; Dunshea, F.R.; Suleria, H.A. Evaluation of the antioxidant potential of brown seaweeds extracted by different solvents and characterization of their phenolic compounds by LC–ESI-QTOF–MS/MS. J. Food Sci. 2023, 88, 3737–3757. [Google Scholar] [CrossRef]
- Ummat, V.; Tiwari, B.K.; Jaiswal, A.K.; Condon, K.; Garcia-Vaquero, M.; O’Doherty, J.; O’Donnell, C.; Rajauria, G. Optimisation of Ultrasound Frequency, Extraction Time and Solvent for the Recovery of Polyphenols, Phlorotannins and Associated Antioxidant Activity from Brown Seaweeds. Mar. Drugs 2020, 18, 250. [Google Scholar] [CrossRef]
- Rajauria, G. Optimization and validation of reverse phase HPLC method for qualitative and quantitative assessment of polyphenols in seaweed. J. Pharm. Biomed. Anal. 2018, 148, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Friedman, M.; Jürgens, H.S. Effect of pH on the Stability of Plant Phenolic Compounds. J. Agric. Food Chem. 2000, 48, 2101–2110. [Google Scholar] [CrossRef] [PubMed]
- Imtiaz, F.; Ahmed, D.; Abdullah, R.H.; Ihsan, S. Green extraction of bioactive compounds from Thuja orientalis leaves using microwave- and ultrasound-assisted extraction and optimization by response surface methodology. Sustain. Chem. Pharm. 2023, 35, 101212. [Google Scholar] [CrossRef]
- Vázquez-Rodríguez, B.; Gutiérrez-Uribe, J.A.; Antunes-Ricardo, M.; Santos-Zea, L.; Cruz-Suárez, L.E. Ultrasound-assisted extraction of phlorotannins and polysaccharides from Silvetia compressa (Phaeophyceae). J. Appl. Phycol. 2020, 32, 1441–1453. [Google Scholar] [CrossRef]
- Živković, J.; Šavikin, K.; Janković, T.; Ćujić, N.; Menković, N. Optimization of ultrasound-assisted extraction of polyphenolic compounds from pomegranate peel using response surface methodology. Sep. Purif. Technol. 2018, 194, 40–47. [Google Scholar] [CrossRef]
- Carrera, C.; Ruiz-Rodríguez, A.; Palma, M.; Barroso, C.G. Ultrasound assisted extraction of phenolic compounds from grapes. Anal. Chim. Acta 2012, 732, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Aquino, G.; Basilicata, M.G.; Crescenzi, C.; Vestuto, V.; Salviati, E.; Cerrato, M.; Ciaglia, T.; Sansone, F.; Pepe, G.; Campiglia, P. Optimization of microwave-assisted extraction of antioxidant compounds from spring onion leaves using Box–Behnken design. Sci. Rep. 2023, 13, 14923. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Zhao, Y.; Wang, L.; Tan, Y.; Shi, Y.; Sedjoah, R.-C.A.-A.; Shao, Y.; Li, L.; Wang, M.; Wan, J. Ultrasound-assisted extraction of bound phenolic compounds from the residue of Apocynum venetum tea and their antioxidant activities. Food Biosci. 2022, 47, 101646. [Google Scholar] [CrossRef]
- Sanou, A.; Konaté, K.; Kabakdé, K.; Dakuyo, R.; Bazié, D.; Hemayoro, S.; Dicko, M.H. Modelling and optimisation of ultrasound-assisted extraction of roselle phenolic compounds using the surface response method. Sci. Rep. 2023, 13, 358. [Google Scholar] [CrossRef] [PubMed]
- Gonzales, G.B.; Smagghe, G.; Raes, K.; Van Camp, J. Combined alkaline hydrolysis and ultrasound-assisted extraction for the release of nonextractable phenolics from cauliflower (Brassica oleracea var. botrytis) waste. J. Agric. Food Chem. 2014, 62, 3371–3376. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Zhang, S.; Wang, H.; Yang, J.; Li, L.; Zhu, J.; Liu, Y. Ultrasound-alkaline combined extraction improves the release of bound polyphenols from pitahaya (Hylocereus undatus ‘Foo-Lon’) peel: Composition, antioxidant activities and enzyme inhibitory activity. Ultrason. Sonochem. 2022, 90, 106213. [Google Scholar] [CrossRef]
- Beuder, S.; Braybrook, S.A. Brown algal cell walls and development. Semin. Cell Dev. Biol. 2023, 134, 103–111. [Google Scholar] [CrossRef]
- Generalić Mekinić, I.; Skroza, D.; Šimat, V.; Hamed, I.; Čagalj, M.; Popović Perković, Z. Phenolic Content of Brown Algae (Pheophyceae) Species: Extraction, Identification, and Quantification. Biomolecules 2019, 9, 244. [Google Scholar] [CrossRef]
- Dang, T.T.; Van Vuong, Q.; Schreider, M.J.; Bowyer, M.C.; Van Altena, I.A.; Scarlett, C.J. Optimisation of ultrasound-assisted extraction conditions for phenolic content and antioxidant activities of the alga Hormosira banksii using response surface methodology. J. Appl. Phycol. 2017, 29, 3161–3173. [Google Scholar] [CrossRef]
- Martínez-López, R.; Tuohy, M.G. Rapid and cost-efficient microplate assay for the accurate quantification of total phenolics in seaweeds. Food Chem. Mol. Sci. 2023, 6, 100166. [Google Scholar] [CrossRef] [PubMed]
- Deniaud-Bouët, E.; Hardouin, K.; Potin, P.; Kloareg, B.; Hervé, C. A review about brown algal cell walls and fucose-containing sulfated polysaccharides: Cell wall context, biomedical properties and key research challenges. Carbohydr. Polym. 2017, 175, 395–408. [Google Scholar] [CrossRef] [PubMed]
- Connan, S.; Stengel, D.B. Impacts of ambient salinity and copper on brown algae: 2. Interactive effects on phenolic pool and assessment of metal binding capacity of phlorotannin. Aquat. Toxicol. 2011, 104, 1–13. [Google Scholar] [CrossRef]
- Jiménez-Escrig, A.; Jiménez-Jiménez, I.; Pulido, R.; Saura-Calixto, F. Antioxidant activity of fresh and processed edible seaweeds. J. Sci. Food Agric. 2001, 81, 530–534. [Google Scholar] [CrossRef]
- Airanthi, M.W.A.; Hosokawa, M.; Miyashita, K. Comparative antioxidant activity of edible Japanese brown seaweeds. J. Food Sci. 2011, 76, C104–C111. [Google Scholar] [CrossRef]
- Budhiyanti, S.A.; Raharjo, S.; Marseno, D.W.; Lelana, I.Y. Antioxidant activity of brown algae Sargassum species extracts from the coastline of java island. Am. J. Agric. Biol. Sci. 2012, 7, 337–346. [Google Scholar] [CrossRef]
- Chakraborty, K.; Maneesh, A.; Makkar, F. Antioxidant Activity of Brown Seaweeds. J. Aquat. Food Prod. Technol. 2017, 26, 406–419. [Google Scholar] [CrossRef]
- Ismail, M.M.; El Zokm, G.M.; Miranda Lopez, J.M. Nutritional, bioactive compounds content, and antioxidant activity of brown seaweeds from the Red Sea. Front. Nutr. 2023, 10, 1210934. [Google Scholar] [CrossRef]
- Afrin, F.; Ahsan, T.; Mondal, M.; Rasul, M.; Afrin, M.; Silva, A.; Yuan, C.; Shah, A. Evaluation of antioxidant and antibacterial activities of some selected seaweeds from Saint Martin’s Island of Bangladesh. Food Chem. Adv. 2023, 3, 100393. [Google Scholar] [CrossRef]
- Kumar, Y.; Singhal, S.; Tarafdar, A.; Pharande, A.; Ganesan, M.; Badgujar, P.C. Ultrasound assisted extraction of selected edible macroalgae: Effect on antioxidant activity and quantitative assessment of polyphenols by liquid chromatography with tandem mass spectrometry (LC-MS/MS). Algal Res. 2020, 52, 102114. [Google Scholar] [CrossRef]
- Agregán, R.; Munekata, P.E.S.; Franco, D.; Dominguez, R.; Carballo, J.; Lorenzo, J.M. Phenolic compounds from three brown seaweed species using LC-DAD–ESI-MS/MS. Food Res. Int. 2017, 99, 979–985. [Google Scholar] [CrossRef] [PubMed]
- Rajauria, G.; Foley, B.; Abu-Ghannam, N. Identification and characterization of phenolic antioxidant compounds from brown Irish seaweed Himanthalia elongata using LC-DAD–ESI-MS/MS. Innov. Food Sci. Emerg. Technol. 2016, 37, 261–268. [Google Scholar] [CrossRef]
- Fernando, I.P.S.; Lee, W.; Ahn, G. Marine algal flavonoids and phlorotannins; an intriguing frontier of biofunctional secondary metabolites. Crit. Rev. Biotechnol. 2022, 42, 23–45. [Google Scholar] [CrossRef] [PubMed]
- Bredsdorff, L.; Nielsen, I.L.F.; Rasmussen, S.E.; Cornett, C.; Barron, D.; Bouisset, F.; Offord, E.; Williamson, G. Absorption, conjugation and excretion of the flavanones, naringenin and hesperetin from α-rhamnosidase-treated orange juice in human subjects. Br. J. Nutr. 2010, 103, 1602–1609. [Google Scholar] [CrossRef] [PubMed]
- Docampo, M.; Olubu, A.; Wang, X.; Pasinetti, G.; Dixon, R.A. Glucuronidated Flavonoids in Neurological Protection: Structural Analysis and Approaches for Chemical and Biological Synthesis. J. Agric. Food Chem. 2017, 65, 7607–7623. [Google Scholar] [CrossRef] [PubMed]
- Mikulski, D.; Molski, M. Quantitative structure–antioxidant activity relationship of trans-resveratrol oligomers, trans-4, 4′-dihydroxystilbene dimer, trans-resveratrol-3-O-glucuronide, glucosides: Trans-piceid, cis-piceid, trans-astringin and trans-resveratrol-4′-O-β-D-glucopyranoside. Eur. J. Med. Chem. 2010, 45, 2366–2380. [Google Scholar] [PubMed]
- Haoujar, I.; Cacciola, F.; Abrini, J.; Mangraviti, D.; Giuffrida, D.; Oulad El Majdoub, Y.; Kounnoun, A.; Miceli, N.; Fernanda Taviano, M.; Mondello, L. The contribution of carotenoids, phenolic compounds, and flavonoids to the antioxidative properties of marine microalgae isolated from Mediterranean Morocco. Molecules 2019, 24, 4037. [Google Scholar] [CrossRef] [PubMed]
- Rajauria, G. In-Vitro Antioxidant Properties of Lipophilic Antioxidant Compounds from 3 Brown Seaweed. Antioxidants 2019, 8, 596. [Google Scholar] [CrossRef]

















| Symbols | Independent Variables | −1 | 0 | 1 |
|---|---|---|---|---|
| Free phenolic extraction | ||||
| X1 | Amplitude (%) | 40 | 60 | 80 |
| X2 | Time (min) | 4 | 6 | 8 |
| X3 | Solvent:solid ratio | 10 | 15 | 20 |
| Y1 | TPC (mg GAE/g) | |||
| Y2 | TPhC (mg PGE/g) | |||
| Y3 | DPPH (mg TE/g) | |||
| Bound phenolic extraction | ||||
| X1 | Amplitude (%) | 40 | 60 | 80 |
| X2 | Time (min) | 6 | 8 | 10 |
| X3 | NaOH concentration (M) | 0.5 | 1.0 | 1.5 |
| Y1 | TPC (mg GAE/g) | |||
| Y2 | TPhC (mg PGE/g) | |||
| Y3 | DPPH (mg TE/g) | |||
| p-Value | Cytospora sp. | D. potatorum | S. fallax | E. radiata | P. comosa | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TPC | TPhC | DPPH | TPC | TPhC | DPPH | TPC | TPhC | DPPH | TPC | TPhC | DPPH | TPC | TPhC | DPPH | |
| Model | 0.01 | 0.00 | <0.0001 | 0.04 | 0.00 | <0.0001 | 0.00 | 0.14 | <0.0001 | 0.00 | 0.01 | <0.0001 | 0.01 | 0.02 | <0.0001 |
| A | 0.31 | 0.27 | 0.27 | 0.11 | 0.72 | 0.95 | 0.90 | 0.17 | 0.93 | 0.14 | 0.11 | 0.01 | 0.12 | 0.59 | 0.08 |
| B | 0.30 | 0.08 | 0.07 | 0.02 | 0.09 | 0.02 | 0.04 | 0.20 | 0.11 | 0.01 | 0.89 | 0.84 | 0.12 | 0.25 | 0.07 |
| C | 0.00 | <0.0001 | <0.0001 | 0.00 | <0.0001 | <0.0001 | <0.0001 | 0.01 | <0.0001 | <0.0001 | 0.00 | <0.0001 | 0.00 | 0.00 | <0.0001 |
| A*B | 0.93 | 0.91 | 0.86 | 0.25 | 0.17 | 0.65 | 0.32 | 0.40 | 0.82 | 0.37 | 0.08 | 0.06 | 0.60 | 0.51 | 0.39 |
| A*C | 0.30 | 0.11 | 0.26 | 0.34 | 0.50 | 0.38 | 0.44 | 0.50 | 0.34 | 0.03 | 0.33 | 0.04 | 0.43 | 0.46 | 0.58 |
| B*C | 0.55 | 0.58 | 0.93 | 0.79 | 0.88 | 0.16 | 0.28 | 0.11 | 0.15 | 0.67 | 0.56 | 0.99 | 0.40 | 0.61 | 0.25 |
| A2 | 0.15 | 0.62 | 0.06 | 0.15 | 0.16 | 0.64 | 0.08 | 0.38 | 0.65 | 0.67 | 0.10 | 0.13 | 0.72 | 0.41 | 0.59 |
| B2 | 0.32 | 0.87 | 0.60 | 0.67 | 0.22 | 0.39 | 0.22 | 0.44 | 0.65 | 0.28 | 0.43 | 0.72 | 0.59 | 0.94 | 0.51 |
| C2 | 0.84 | 0.05 | 0.03 | 0.14 | 0.40 | 0.02 | 0.59 | 0.93 | 0.00 | 0.25 | 0.40 | 0.00 | 0.50 | 0.48 | 0.00 |
| Lack of fit | 0.28 | 0.76 | 0.14 | 0.20 | 0.46 | 0.74 | 0.51 | 0.59 | 0.68 | 0.54 | 0.28 | 0.39 | 0.22 | 0.79 | 0.74 |
| R2 | 0.95 | 0.99 | 1.00 | 0.91 | 0.99 | 1.00 | 0.99 | 0.83 | 1.00 | 0.98 | 0.95 | 1.00 | 0.89 | 0.87 | 1.00 |
| Adjusted R2 | 0.93 | 0.97 | 0.99 | 0.75 | 0.96 | 0.99 | 0.96 | 0.53 | 1.00 | 0.94 | 0.87 | 1.00 | 0.68 | 0.63 | 1.00 |
| Adeq. Precision | 9.33 | 20.03 | 32.15 | 8.40 | 18.68 | 39.97 | 21.10 | 6.21 | 54.10 | 15.93 | 9.69 | 72.18 | 6.91 | 6.82 | 48.20 |
| p-Value | Cytospora sp. | D. potatorum | S. fallax | E. radiata | P. comosa | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TPC | TPhC | DPPH | TPC | TPhC | DPPH | TPC | TPhC | DPPH | TPC | TPhC | DPPH | TPC | TPhC | DPPH | |
| Model | 0.01 | 0.01 | 0.05 | 0.01 | 0.01 | 0.03 | 0.07 | 0.11 | 0.15 | 0.03 | 0.04 | 0.02 | 0.05 | 0.11 | 0.07 |
| A | 0.20 | 0.06 | 0.87 | 0.77 | 0.76 | 0.61 | 0.10 | 0.23 | 0.28 | 0.56 | 0.91 | 0.62 | 0.07 | 0.11 | 0.09 |
| B | 0.03 | 0.02 | 0.03 | 0.77 | 0.96 | 0.72 | 0.98 | 0.78 | 0.58 | 0.63 | 0.48 | 0.29 | 0.60 | 0.63 | 0.62 |
| C | 0.75 | 0.00 | 0.92 | 0.00 | 0.00 | 0.00 | 0.07 | 0.05 | 0.06 | 0.00 | 0.00 | 0.00 | 0.12 | 0.08 | 0.06 |
| A*B | 0.01 | 0.21 | 0.05 | 0.03 | 0.07 | 0.14 | 0.20 | 0.19 | 0.18 | 0.39 | 0.62 | 0.41 | 0.11 | 0.23 | 0.15 |
| A*C | 0.09 | 0.15 | 0.51 | 0.86 | 0.87 | 0.50 | 0.44 | 0.62 | 0.76 | 0.29 | 0.34 | 0.62 | 0.23 | 0.34 | 0.19 |
| B*C | 0.32 | 0.09 | 0.50 | 0.13 | 0.12 | 0.12 | 0.57 | 0.39 | 0.13 | 0.05 | 0.07 | 0.05 | 0.49 | 0.57 | 0.86 |
| A2 | 0.00 | 0.00 | 0.01 | 0.05 | 0.11 | 0.27 | 0.14 | 0.20 | 0.21 | 0.87 | 0.53 | 0.55 | 0.03 | 0.15 | 0.09 |
| B2 | 0.51 | 0.62 | 0.90 | 0.01 | 0.02 | 0.04 | 0.01 | 0.02 | 0.05 | 0.05 | 0.02 | 0.01 | 0.01 | 0.02 | 0.01 |
| C2 | 0.02 | 0.30 | 0.06 | 0.47 | 0.38 | 0.96 | 0.18 | 0.29 | 0.60 | 0.07 | 0.34 | 0.06 | 0.67 | 0.65 | 0.68 |
| Lack of fit | 0.27 | 0.22 | 0.27 | 0.10 | 0.32 | 0.26 | 0.27 | 0.15 | 0.09 | 0.41 | 0.38 | 0.57 | 0.42 | 0.25 | 0.36 |
| R2 | 0.95 | 0.95 | 0.90 | 0.89 | 0.90 | 0.86 | 0.80 | 0.80 | 0.84 | 0.86 | 0.84 | 0.86 | 0.83 | 0.77 | 0.81 |
| Adjusted R2 | 0.85 | 0.85 | 0.72 | 0.76 | 0.76 | 0.67 | 0.74 | 0.78 | 0.76 | 0.68 | 0.75 | 0.68 | 0.78 | 0.68 | 0.76 |
| Adeq. Precision | 8.55 | 10.25 | 6.33 | 7.28 | 7.98 | 7.24 | 5.30 | 4.84 | 4.97 | 7.88 | 7.44 | 8.07 | 6.43 | 5.54 | 6.97 |
| Samples | Optimal Extraction Conditions | Values | TPC (mg GAE/g) | TPhC (mg PGE/g) | DPPH (mg TE/g) |
|---|---|---|---|---|---|
| Free Phenolics | |||||
| Cytospora sp. | Amplitude 52% | Predicted | 15.57 | 2.34 | 60.64 |
| Time 8 min | Experimental | 14.64 ± 1.12 | 2.42 ± 0.23 | 60.67 ± 0.17 | |
| Solvent–solid ratio 20 | 95% CI | 12.21–19.34 | 2.13–2.55 | 56.40–65.03 | |
| Durvilleae potatorum | Amplitude 51% | Predicted | 3.92 | 2.48 | 35.91 |
| Time 8 min | Experimental | 4.01 ± 0.29 | 0.43 ± 0.02 | 35.26 ± 0.08 | |
| Solvent–solid ratio 20 | 95% CI | 3.08–4.77 | 2.08–2.88 | 33.18–38.68 | |
| Sargassum fallax | Amplitude 57% | Predicted | 21.46 | 1.20 | 73.07 |
| Time 8 min | Experimental | 20.32 ± 0.41 | 1.28 ± 0.02 | 73.66 ± 0.51 | |
| Solvent–solid ratio 20 | 95% CI | 19.76–23.16 | 18.87–24.05 | 68.90–77.24 | |
| Ecklonia radiata | Amplitude 80% | Predicted | 11.51 | 2.76 | 67.46 |
| Time 8 min | Experimental | 11.6 ± 0.57 | 1.01 ± 0.02 | 66.84 ± 0.34 | |
| Solvent–solid ratio 20 | 95% CI | 10.16–12.87 | 2.13–3.39 | 64.19–70.73 | |
| Phyllospora comosa | Amplitude 80% | Predicted | 8.33 | 0.83 | 65.69 |
| Time 8 min | Experimental | 8.15 ± 0.20 | 0.94 ± 0.01 | 64.38 ±0.80 | |
| Solvent–solid ratio 20 | 95% CI | 5.98–10.67 | 0.61–1.05 | 60.89–70.48 | |
| Bound Phenolics | |||||
| Cytospora sp. | Amplitude 40% | Predicted | 4.45 | 1.52 | 22.84 |
| Time 10 min | Experimental | 4.49 ± 0.02 | 1.46 ± 0.04 | 20.15 ± 0.04 | |
| NaOH 1.5 M | 95% CI | 4.21–4.67 | 1.42–1.61 | 21.60–23.90 | |
| Durvilleae potatorum | Amplitude 80% | Predicted | 2.54 | 0.70 | 15.62 |
| Time 6 min | Experimental | 2.97 ± 0.02 | 0.76 ± 0.02 | 14.51 ± 0.04 | |
| NaOH 0.5 M | 95% CI | 2.31–2.98 | 0.67–0.74 | 14.8–16.4 | |
| Sargassum fallax | Amplitude 40% | Predicted | 4.62 | 1.69 | 21.20 |
| Time 10 min | Experimental | 4.34 ± 0.01 | 1.62 ± 0.01 | 19.96 ± 0.12 | |
| NaOH 1.5 M | 95% CI | 4.39–8.65 | 1.61–1.77 | 19.84–22.26 | |
| Ecklonia radiata | Amplitude 40% | Predicted | 1.42 | 0.38 | 4.26 |
| Time 8 min | Experimental | 1.41 ± 0.00 | 0.48 ± 0.01 | 4.68 ± 0.05 | |
| NaOH 1.5 M | 95% CI | 1.35–1.49 | 0.36–0.40 | 4.05–4.47 | |
| Phyllospora comosa | Amplitude 42% | Predicted | 3.98 | 0.68 | 9.56 |
| Time 8 min | Experimental | 4.24 ± 0.01 | 0.71 ± 0.01 | 9.91 ± 0.07 | |
| NaOH 1.5 M | 95% CI | 3.78–4. 25 | 0.65–0.72 | 9.08–10.04 | |
| Samples | TPC (mg GAE/g) | TFC (mg QE/g) | TCT (mg CE/g) | TPhC (mg PGE/g) | DPPH (mg TE/g) | ABTS (mg TE/g) | FRAP (mg TE/g) | PRAC (mg TE/g) | FICA (mg EDTA-E/g) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Free Phenolics | Conventional Extraction | |||||||||
| Cytospora sp. | 10.62 ± 0.24 Aa | 0.34 ± 0.02 Aa | 2.39 ± 0.51 Aa | 1.22 ± 0.04 Aa | 39.97 ± 0.64 Aa | 44.86 ± 1.03 Aa | 19.91 ± 0.35 Aa | 22.92 ± 0.53 Aa | 3.32 ± 0.14 Aa | |
| Durvillaea potatorum | 3.61 ± 0.01 Ab | 0.03 ± 0.00 Ab | - | 0.15 ± 0.00 Ab | 20.85 ± 0.26 Ab | 20.27 ± 0.19 Ab | 0.98 ± 0.04 Ab | 0.36 ± 0.02 Ab | 3.17 ± 0.11 Aab | |
| Sargassum fallax | 17.43 ± 0.02 Ac | 0.41 ± 0.02 Ac | - | 0.74 ± 0.02 Ac | 49.97 ± 1.14 Ac | 50.40 ± 1.80 Ac | 24.52 ± 0.77 Ac | 46.37 ± 0.77 Ac | 3.05 ± 0.01 Ab | |
| Ecklonia radiata | 8.95 ± 0.06 Ad | 0.01 ± 0.00 Ab | 6.23 ± 0.61 Ab | 2.03 ± 0.03 Ad | 32.80 ± 0.37 Ad | 46.87 ± 0.79 Aa | 15.19 ± 0.43 Ad | 3.11 ± 0.08 Ad | 2.74 ± 0.04 Ac | |
| Phyllospora comosa | 4.03 ± 0.09 Ae | 0.03 ± 0.00 Ab | - | 0.35 ± 0.01 Ae | 22.58 ± 0.36 Ae | 27.98 ± 0.78 Ad | 5.43 ± 0.19 Ae | 3.12 ± 0.06 Ad | 3.15 ± 0.05 Aab | |
| Ultrasonic Extraction | ||||||||||
| Cytospora sp. | 14.64 ± 1.12 Ba | 0.96 ± 0.05 Ba | 4.35 ± 0.47 Ba | 2.42 ± 0.23 Ba | 60.67 ± 0.17 Ba | 88.08 ± 1.55 Ba | 29.21 ± 0.46 Ba | 39.60 ± 0.66 Ba | 11.78 ± 0.14 Ba | |
| Durvillaea potatorum | 4.01 ± 0.29 Ab | 0.43 ± 0.02 Bb | 3.48 ± 0.21 b | 2.26 ± 0.25 Bb | 35.26 ± 0.08 Bb | 31.44 ± 0.68 Bb | 1.17 ± 0.04 Bb | 0.84 ± 0.04 Bb | 7.52 ± 0.08 Bb | |
| Sargassum fallax | 20.32 ± 0.41 Bc | 1.28 ± 0.02 Bc | - | 1.16 ± 0.29 Ac | 73.66 ± 0.51 Bc | 86.88 ± 0.40 Ba | 33.70 ± 0.68 Bc | 60.68 ± 0.05 Bc | 6.76 ± 0.06 Bc | |
| Ecklonia radiata | 11.6 ± 0.57 Bd | 1.01 ± 0.02 Bd | 7.80 ± 0.18 Bc | 2.64 ± 0.15 Bb | 66.84 ± 0.34 Bd | 80.38 ± 0.39 Bc | 17.70 ± 0.56 Bd | 5.53 ± 0.21 Bd | 5.95 ± 0.17 Bd | |
| Phyllospora comosa | 8.15 ± 0.20 Be | 0.94 ± 0.01 Ba | 0.38 ± 0.12 d | 0.81 ± 0.04 Bc | 64.38 ± 0.80 Be | 39.95 ± 0.90 Bd | 9.60 ± 0.04 Be | 3.72 ± 0.12 Be | 5.42 ± 0.11 Be | |
| Bound Phenolics | Conventional Extraction | |||||||||
| Cytospora sp. | 1.25 ± 0.04 Aa | 0.10 ± 0.01 Aab | - | 0.18 ± 0.00 Aa | 3.77 ± 0.11 Aa | 7.63 ± 0.11 Aa | 3.20 ± 0.08 Aa | 3.51 ± 0.02 Aa | 1.00 ± 0.01 Aa | |
| Durvillaea potatorum | 2.40 ± 0.04 Ab | 0.01 ± 0.00 Ac | 5.54 ± 0.02 Aa | 0.42 ± 0.01 Ab | 5.18 ± 0.12 Ab | 8.71 ± 0.20 Ab | 2.09 ± 0.05 Ab | 0.21 ± 0.01 Ab | 0.60 ± 0.01 Ab | |
| Sargassum fallax | 0.66 ± 0.02 Ac | 0.13 ± 0.02 Ad | - | 0.08 ± 0.00 Ac | 2.97 ± 0.01 Ac | 4.00 ± 0.00 Ac | 2.07 ± 0.01 Ab | 0.43 ± 0.05 Ac | 0.56 ± 0.00 Ac | |
| Ecklonia radiata | 0.46 ± 0.01 Ad | 0.10 ± 0.01 Aa | - | 0.02 ± 0.00 Ad | 2.54 ± 0.05 Ad | 3.56 ± 0.06 Ad | 1.02 ± 0.04 Ac | 1.16 ± 0.01 Ad | 0.50 ± 0.01 Ad | |
| Phyllospora comosa | 0.30 ± 0.01 Ae | 0.10 ± 0.00 Ab | 0.19 ± 0.06 Ab | 0.15 ± 0.01 Ae | 1.83 ± 0.04 Ae | 2.75 ± 0.04 Ae | 1.32 ± 0.02 Ad | 1.25 ± 0.08 Ae | 0.46 ± 0.01 Ae | |
| Ultrasonic Extraction | ||||||||||
| Cytospora sp. | 4.49 ± 0.02 Ba | 0.25 ± 0.09 Ba | - | 1.46 ± 0.04 Ba | 20.15 ± 0.04 Ba | 21.42 ± 0.86 Ba | 8.44 ± 0.18 Ba | 10.4 ± 0.19 Ba | 1.42 ± 0.03 Ba | |
| Durvillaea potatorum | 2.97 ± 0.02 Bb | 0.07 ± 0.00 Bb | 6.91 ± 0.09 Aa | 0.76 ± 0.02 Bb | 14.51 ± 0.04 Bb | 17.96 ± 0.09 Bb | 2.25 ± 0.08 Bb | 6.15 ± 0.15 Bb | 0.92 ± 0.01 Bb | |
| Sargassum fallax | 4.34 ± 0.01 Bc | 0.00 ± 0.00 Bc | 1.63 ± 0.38 bc | 1.62 ± 0.01 Bc | 19.96 ± 0.12 Ba | 6.47 ± 0.18 Bc | 2.79 ± 0.01 Bc | 0.52 ± 2.62 Bc | 1.02 ± 0.03 Bc | |
| Ecklonia radiata | 1.41 ± 0.00 Bd | 0.08 ± 0.01 Bd | 3.15 ± 0.78 b | 0.48 ± 0.01 Bd | 4.68 ± 0.05 Bc | 5.65 ± 0.20 Bc | 1.16 ± 0.04 Bd | 2.12 ± 0.54 Bd | 1.10 ± 0.01 Bd | |
| Phyllospora comosa | 4.24 ± 0.01 Be | 0.04 ± 0.01 Be | 0.43 ± 0.08 Ac | 0.71 ± 0.01 Bb | 9.91 ± 0.07 Bd | 7.73 ± 0.34 Bd | 1.50 ± 0.03 Be | 1.95 ± 0.19 Bc | 1.01 ± 0.03 Bc | |
| No. | Proposed Compounds | Molecular Formula | RT (min) | Ionization (ESI+/ESI−) | Molecular Weight | Theoretical (m/z) | Observed (m/z) | Error (ppm) | Product Ion (m/z) | Sample |
|---|---|---|---|---|---|---|---|---|---|---|
| PHENOLIC ACIDS | ||||||||||
| Hydroxycinnamic acid | ||||||||||
| 1 | 1,2,2′-Triferuloylgentiobiose | C42H46O20 | 7.766 | [M − H]− | 870.2613 | 869.2540 | 869.2526 | −1.6 | 693, 517 | * CFEC, UFEC, CBEC |
| 2 | p-Coumaroyl malic acid | C13H12O7 | 64.185 | [M − H]− | 280.0575 | 279.0502 | 279.0509 | 2.5 | 163, 119 | * UFCY, UBCY, CBDU, UFPH |
| 3 | 1-Sinapoyl-2,2′-diferuloylgentiobiose | C43H48O21 | 67.219 | [M − H]− | 900.2671 | 899.2598 | 899.2617 | 2.1 | 613, 201 | UFEC |
| Hydroxybenzoic acid | ||||||||||
| 4 | 3,4-O-Dimethylgallic acid | C9H10O5 | 40.416 | [M + H]+ | 198.0537 | 199.061 | 199.061 | 0.0 | 153, 139, 125, 111 | * CBDU, CBSA, CBPH, CFCY, CFPH, UBCY, UBDU, UBSA, UBPH |
| 5 | 4-O-Methylgallic acid | C8H8O5 | 55.241 | [M + H]+ | 184.0381 | 185.0454 | 185.0452 | −1.1 | 170, 141 | * CBSA, CFDU, UBCY, UBDU |
| FLAVONOIDS | ||||||||||
| Flavanols | ||||||||||
| 6 | Quercetin 3-O-xylosyl-glucuronide | C26H26O17 | 13.846 | [M + H]+ | 610.1123 | 611.1196 | 611.1223 | 4.4 | 479, 303, 285, 239 | * UFDU, CFDU, CFSA, UFSA |
| 7 | Quercetin 3′-sulfate | C15H10O10S | 14.006 | [M − H]− | 381.9973 | 380.9900 | 380.9891 | −2.4 | 301 | * CBEC, CBDU |
| 8 | Prodelphinidin dimer B3 | C30H26O14 | 54.815 | [M + H]+ | 610.13 | 611.1373 | 611.1384 | 1.8 | 469, 311, 291 | * UFCY, CFCY, UFPH |
| 9 | Spinacetin 3-O-glucosyl-(1->6)-glucoside | C29H34O18 | 62.464 | [M − H]− | 670.1743 | 669.1670 | 669.1655 | −2.2 | 609, 301 | CFDU |
| Anthocyanins | ||||||||||
| 10 | Cyanidin 3-O-(6″-p-coumaroyl-glucoside) | C30H27O13 | 55.998 | [M + H]+ | 595.1474 | 596.1547 | 596.1554 | 1.2 | 287 | CFPH |
| 11 | Cyanidin 3-O-diglucoside-5-O-glucoside | C33H41O21 | 57.989 | [M + H]+ | 773.2171 | 774.2244 | 774.2246 | 0.3 | 610, 464 | UFCY |
| 12 | Peonidin 3-O-diglucoside-5-O-glucoside | C34H43O21 | 63.639 | [M + H]+ | 787.2335 | 788.2408 | 788.2445 | 4.7 | 625, 478, 317 | UBPH |
| Flavanones | ||||||||||
| 13 | Neohesperidin | C28H34O15 | 13.784 | [M + H]+ | 610.1908 | 611.1981 | 611.1985 | 0.7 | CFPH | |
| 14 | Hesperetin 5,7-O-diglucuronide | C28H30O18 | 63.386 | [M − H]− | 654.1372 | 653.1299 | 653.132 | 3.2 | 447, 301, 286, 242 | * CFEC, CFDU |
| Flavones | ||||||||||
| 15 | Nobiletin | C21H22O8 | 67.568 | [M + H]+ | 402.1323 | 403.1396 | 403.1393 | −0.7 | 359 | * CFCY, UFCY |
| Isoflavonoids | ||||||||||
| 16 | 6″-O-Acetylglycitin | C24H24O11 | 13.841 | ** [M + H]+ | 488.1326 | 487.1253 | 487.1259 | 1.2 | 285, 270 | * CFCY, CFSA, CFSA, CFPH, UFCY, UFSA, UFPH |
| OTHER POLYPHENOLS | ||||||||||
| Hydroxycoumarins | ||||||||||
| 17 | Coumarin | C9H6O2 | 13.576 | [M + H]+ | 146.0379 | 147.0452 | 147.0451 | −0.7 | 103, 91 | UFCY |
| 18 | Esculin | C15H16O9 | 14.058 | [M + H]+ | 340.0813 | 341.0886 | 341.0882 | −1.2 | 179, 151 | CBPH |
| Hydroxyphenylpropenes | ||||||||||
| 19 | Eugenol | C10H12O2 | 65.708 | [M + H]+ | 164.0845 | 165.0918 | 165.0916 | −1.2 | 153 | * CFPH, CBEC, UBCY, UBSA, UBEC, UBPH, UFPH |
| Hydroxybenzaldehydes | ||||||||||
| 20 | p-Anisaldehyde | C8H8O2 | 63.263 | [M + H]+ | 136.0528 | 137.0601 | 137.0602 | 0.7 | 122, 109 | * CFEC, CBSA, CBEC, CFPH |
| Phenolic terpenes | ||||||||||
| 21 | Epirosmanol | C20H26O5 | 59.444 | [M + H]+ | 346.1793 | 347.1866 | 347.1852 | −4.0 | 253 | * CFEC, CBEC, CBPH, UBEC, UFEC |
| Curcuminoids | ||||||||||
| 22 | Bisdemethoxycurcumin | C19H16O4 | 13.874 | [M + H]+ | 308.1051 | 309.1124 | 309.1122 | −0.6 | 291, 263 | * CFDU, CFPH |
| LIGNANS | ||||||||||
| 23 | Enterolactone | C18H18O4 | 65.845 | [M + H]+ | 298.1184 | 299.1257 | 299.1257 | 0.0 | 281, 187, 165 | CFEC |
| 24 | 7-Oxomatairesinol | C20H20O7 | 57.618 | [M + H]+ | 372.1212 | 373.1285 | 373.1276 | −2.4 | 358, 343, 328, 325 | CBPH |
| 25 | Schisandrin | C24H32O7 | 60.864 | [M + H]+ | 432.2139 | 433.2212 | 433.2205 | −1.6 | 415, 361 | * CBEC, CBCY, CBPH |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, Z.J.; Xie, C.; Duan, X.; Ng, K.; Suleria, H.A.R. Optimization of Ultrasonic Extraction Parameters for the Recovery of Phenolic Compounds in Brown Seaweed: Comparison with Conventional Techniques. Antioxidants 2024, 13, 409. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox13040409
Lee ZJ, Xie C, Duan X, Ng K, Suleria HAR. Optimization of Ultrasonic Extraction Parameters for the Recovery of Phenolic Compounds in Brown Seaweed: Comparison with Conventional Techniques. Antioxidants. 2024; 13(4):409. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox13040409
Chicago/Turabian StyleLee, Zu Jia, Cundong Xie, Xinyu Duan, Ken Ng, and Hafiz A. R. Suleria. 2024. "Optimization of Ultrasonic Extraction Parameters for the Recovery of Phenolic Compounds in Brown Seaweed: Comparison with Conventional Techniques" Antioxidants 13, no. 4: 409. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox13040409