Association between the blaCTX-M-14-harboring Escherichia coli Isolated from Weasels and Domestic Animals Reared on a University Campus
Abstract
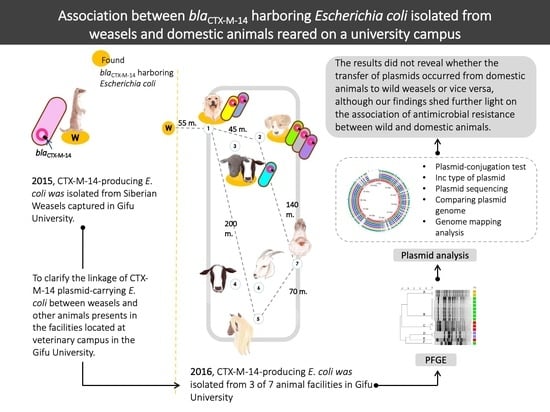
:1. Introduction
2. Results
2.1. The Infection Rate and Antimicrobial Susceptibility Profiles of ESBL-Producing E. coli in Animal Facilities and Wild Siberian Weasels
| Animal Facility (Number of Sampling Location a) | Collection Period | Animal Species (Number of Samples) | Number of CTX-M-14- Positive Samples (Prevalence, %) |
|---|---|---|---|
| Dog facility 1 (1) | June | Dog (11) | 8 (72.7) b |
| Dog facility 2 (2) | December | Dog (10) | 4 (40) b |
| Cattle facility (3) | October | Dairy cow (5) Beef cow (2) | 2 (28.6) |
| Dairy cow facility (4) | October | Dairy cow (18) | 0 (0) |
| Pony facility (5) | June | Pony (4) | 0 (0) |
| Goat facility (6) | August | Goat (14) | 0 (0) |
| Laying hen facility (7) | September | Laying hen (5) | 0 (0) |
| Total number of samples | 69 | 14 (20.3) |
2.2. PFGE Analysis
2.3. CTX-M-14 Plasmid Analysis
2.3.1. Genome Comparisons of the CTX-M-14 Plasmids
2.3.2. Relatedness between Six CTX-M-14 Plasmids and Their Interrelatedness with Highly Similar Plasmids
3. Discussion
4. Materials and Methods
4.1. Sampling and Bacterial Isolation
4.1.1. Samples from Siberian Weasels
4.1.2. Samples from Domestic Animals from Animal Facilities
4.2. Ethics Statement
4.3. Antimicrobial Susceptibility Testing
4.4. β-Lactamase Gene Identification
4.5. PFGE Analysis
4.6. Selected Isolates for Plasmid Sequencing
4.7. Analysis of Plasmid Sequences
4.7.1. CTX-M-14 Plasmid Similarity Analysis
4.7.2. Relatedness among All Plasmids Used in this Study and Interrelatedness between the Highly Similar Plasmids
4.7.3. Nucleotide Sequence Accession Numbers
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Woolhouse, M.; Ward, M.; van Bunnik, B.; Farrar, J. Antimicrobial resistance in humans, livestock and the wider environment. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20140083. [Google Scholar] [CrossRef] [PubMed]
- Ho, P.L.; Lo, W.U.; Lai, E.L.; Law, P.Y.; Leung, S.M.; Wang, Y.; Chow, K.H. Clonal diversity of CTX-M-producing, multidrug-resistant Escherichia coli from rodents. J. Med. Microbiol. 2015, 64, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, I.; Obi, T.; Sakemi, Y.; Nakayama, A.; Miyazaki, K.; Ogura, G.; Tamaki, M.; Oka, T.; Takase, K.; Miyamoto, A.; et al. The prevalence of antimicrobial-resistant Escherichia coli in two species of invasive alien mammals in Japan. J. Vet. Med. Sci. 2011, 73, 1067–1070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Ma, Z.B.; Zeng, Z.L.; Yang, X.W.; Huang, Y.; Liu, J.H. The role of wildlife (wild birds) in the global transmission of antimicrobial resistance genes. Zool. Res. 2017, 38, 55–80. [Google Scholar] [CrossRef] [Green Version]
- Seiffert, S.N.; Hilty, M.; Perreten, V.; Endimiani, A. Extended-spectrum cephalosporin-resistant gram-negative organisms in livestock: An emerging problem for human health? Drug Resist. Updat. 2013, 16, 22–45. [Google Scholar] [CrossRef]
- Hiki, M.; Usui, M.; Kojima, A.; Ozawa, M.; Ishii, Y.; Asai, T. Diversity of plasmid replicons encoding the bla(CMY-2) gene in broad-spectrum cephalosporin-resistant Escherichia coli from livestock animals in Japan. Foodborne Pathog. Dis. 2013, 10, 243–249. [Google Scholar] [CrossRef]
- Okubo, T.; Sato, T.; Yokota, S.; Usui, M.; Tamura, Y. Comparison of broad-spectrum cephalosporin-resistant Escherichia coli isolated from dogs and humans in Hokkaido, Japan. J. Infect. Chemother. 2014, 20, 243–249. [Google Scholar] [CrossRef]
- Norizuki, C.; Kawamura, K.; Wachino, J.I.; Suzuki, M.; Nagano, N.; Kondo, T.; Arakawa, Y. Detection of Escherichia coli producing CTX-M-1-group extended-spectrum beta-lactamases from pigs in Aichi prefecture, Japan, between 2015 and 2016. Jpn. J. Infect. Dis. 2018, 71, 33–38. [Google Scholar] [CrossRef] [Green Version]
- Carattoli, A.; Bertini, A.; Villa, L.; Falbo, V.; Hopkins, K.L.; Threlfall, E.J. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 2005, 63, 219–228. [Google Scholar] [CrossRef]
- Hulter, N.; Ilhan, J.; Wein, T.; Kadibalban, A.S.; Hammerschmidt, K.; Dagan, T. An evolutionary perspective on plasmid lifestyle modes. Curr. Opin. Microbiol. 2017, 38, 74–80. [Google Scholar] [CrossRef]
- Norizuki, C.; Wachino, J.I.; Suzuki, M.; Kawamura, K.; Nagano, N.; Kimura, K.; Arakawa, Y. Specific blaCTX-M-8/IncI1 plasmid transfer among genetically diverse Escherichia coli isolates between humans and chickens. Antimicrob. Agents Chemother. 2017, 61, 17. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, H. Mustela sibirica Pallas. In The Wild Mammals of Japan; Ohdachi, S.D., Ishibashi, Y., Iwasa, M.A., Fukui, S.T., Eds.; Shoukadoh: Kyoto, Japan, 2009; pp. 250–251. ISBN 13 978-4879746269. [Google Scholar]
- Moodley, A.; Guardabassi, L. Transmission of IncN plasmids carrying blaCTX-M-1 between commensal Escherichia coli in pigs and farm workers. Antimicrob. Agents Chemother. 2009, 53, 1709–1711. [Google Scholar] [CrossRef] [Green Version]
- Schink, A.K.; Kadlec, K.; Schwarz, S. Analysis of bla(CTX-M)-carrying plasmids from Escherichia coli isolates collected in the BfT-GermVet study. Appl. Environ. Microbiol. 2011, 77, 7142–7146. [Google Scholar] [CrossRef] [Green Version]
- Kotrba, P.; Inui, M.; Yukawa, H. Bacterial phosphotransferase system (PTS) in carbohydrate uptake and control of carbon metabolism. J. Biosci. Bioeng. 2001, 92, 502–517. [Google Scholar] [CrossRef]
- Gyohda, A.; Furuya, N.; Ishiwa, A.; Zhu, S.; Komano, T. Structure and function of the shufflon in plasmid R64. Adv. Biophys. 2004, 38, 183–213. [Google Scholar] [CrossRef]
- Sekizuka, T.; Kawanishi, M.; Ohnishi, M.; Shima, A.; Kato, K.; Yamashita, A.; Matsui, M.; Suzuki, S.; Kuroda, M. Elucidation of quantitative structural diversity of remarkable rearrangement regions, shufflons, in IncI2 plasmids. Sci. Rep. 2017, 7, 928. [Google Scholar] [CrossRef]
- Hall, J.P.J.; Brockhurst, M.A.; Dytham, C.; Harrison, E. The evolution of plasmid stability: Are infectious transmission and compensatory evolution competing evolutionary trajectories? Plasmid 2017, 91, 90–95. [Google Scholar] [CrossRef]
- Asai, T.; Usui, M.; Sugiyama, M.; Izumi, K.; Ikeda, T.; Andoh, M. Antimicrobial susceptibility of Escherichia coli isolates obtained from wild mammals between 2013 and 2017 in Japan. J. Vet. Med. Sci. 2020, 3, 345–349. [Google Scholar] [CrossRef] [Green Version]
- CLSI: Performance Standard for Antimicrobial Susceptibility Testing, 26th ed.; CLSI Document M100-S20; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2016.
- EUCAST: Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 9.0. European Committee on Antimicrobial Susceptibility Testing. 2019. Available online: http://www.eucast.org (accessed on 13 April 2021).
- Luzzaro, F.; Mantengoli, E.; Perilli, M.; Lombardi, G.; Orlandi, V.; Orsatti, A.; Amicosante, G.; Rossolini, G.M.; Toniolo, A. Dynamics of a nosocomial outbreak of multidrug-resistant Pseudomonas aeruginosa producing the PER-1 extended-spectrum beta-lactamase. J. Clin. Microbiol. 2001, 39, 1865–1870. [Google Scholar] [CrossRef] [Green Version]
- Dallenne, C.; Costa, A.D.; Decre, D.; Favier, C.; Arlet, G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef] [Green Version]
- Kojima, A.; Ishii, Y.; Ishihara, K.; Esaki, H.; Asai, T.; Oda, C.; Tamura, Y.; Takahashi, T.; Yamaguchi, K. Extended-spectrum-beta-lactamase-producing Escherichia coli strains isolated from farm animals from 1999 to 2002: Report from the Japanese Veterinary Antimicrobial Resistance Monitoring Program. Antimicrob. Agents Chemother. 2005, 49, 3533–3537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Usui, M.; Hiki, M.; Murakami, K.; Ozawa, M.; Nagai, H.; Asai, T. Evaluation of transferability of R-plasmid in bacteriocin-producing donors to bacteriocin-resistant recipients. Jpn. J. Infect. Dis. 2012, 65, 252–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marasini, D.; Fakhr, M.K. Exploring PFGE for detecting large plasmids in Campylobacter jejuni and Campylobacter coli isolated from various retail meats. Pathogens 2014, 3, 833–844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coil, D.; Jospin, G.; Darling, A.E. A5-miseq: An updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 2015, 31, 587–589. [Google Scholar] [CrossRef]
- Richter, M.; Rossello-Mora, R.; Glockner, O.F.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef]
- Petkau, A.; Stuart-Edwards, M.; Stothard, P.; Domselaar, V.G. Interactive microbial genome visualization with GView. Bioinformatics 2010, 26, 3125–3126. [Google Scholar] [CrossRef]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [Green Version]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]





| Animal Origin or Facility | Donor | Transconjugant (TC) | ||||||
|---|---|---|---|---|---|---|---|---|
| Sample ID a | β-Lactamase | Non-β-Lactam-Resistance Profile b | Plasmid ID | β-Lactamase | Non-β-Lactam-Resistance Profile | Replicon Type | Plasmid Size (bp) c | |
| Siberian weasel | MS7 | CTX-M-14 | Susceptible | p130MS | CTX-M-14 | Susceptible | IncI1 | 98,904 |
| Cattle facility (CF) | BC6 | CTX-M-14 | Susceptible | p105CF | CTX-M-14 | Susceptible | IncI1 | 99,001 |
| Dog facility 1 (DF1) | D11 | CTX-M-14, TEM-1 | TET, NAL, CIP, CHL | p74DF1 | CTX-M-14 | Susceptible | IncI1 | 99,262 |
| D12 | CTX-M-14 | Susceptible | p80DF1 | CTX-M-14 | Susceptible | IncI1 | 98,904 | |
| Dog facility 2 (DF2) | D23 | CTX-M-14 | Susceptible | p116DF2 | CTX-M-14 | Susceptible | IncI1 | 99,067 |
| D27 | CTX-M-14, TEM-1 | GEN, KAN, SXT | p123DF2 | CTX-M-14 | Susceptible | IncI1 | 105,498 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yossapol, M.; Yamamoto, M.; Sugiyama, M.; Odoi, J.O.; Omatsu, T.; Mizutani, T.; Ohya, K.; Asai, T. Association between the blaCTX-M-14-harboring Escherichia coli Isolated from Weasels and Domestic Animals Reared on a University Campus. Antibiotics 2021, 10, 432. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10040432
Yossapol M, Yamamoto M, Sugiyama M, Odoi JO, Omatsu T, Mizutani T, Ohya K, Asai T. Association between the blaCTX-M-14-harboring Escherichia coli Isolated from Weasels and Domestic Animals Reared on a University Campus. Antibiotics. 2021; 10(4):432. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10040432
Chicago/Turabian StyleYossapol, Montira, Miku Yamamoto, Michiyo Sugiyama, Justice Opare Odoi, Tsutomu Omatsu, Tetsuya Mizutani, Kenji Ohya, and Tetsuo Asai. 2021. "Association between the blaCTX-M-14-harboring Escherichia coli Isolated from Weasels and Domestic Animals Reared on a University Campus" Antibiotics 10, no. 4: 432. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10040432