Optical DNA Mapping of Plasmids Reveals Clonal Spread of Carbapenem-Resistant Klebsiella pneumoniae in a Large Thai Hospital
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Isolates
2.2. DNA Extraction and Whole Genome Sequencing
2.3. Plasmid Extraction
2.4. Optical DNA Mapping (ODM)
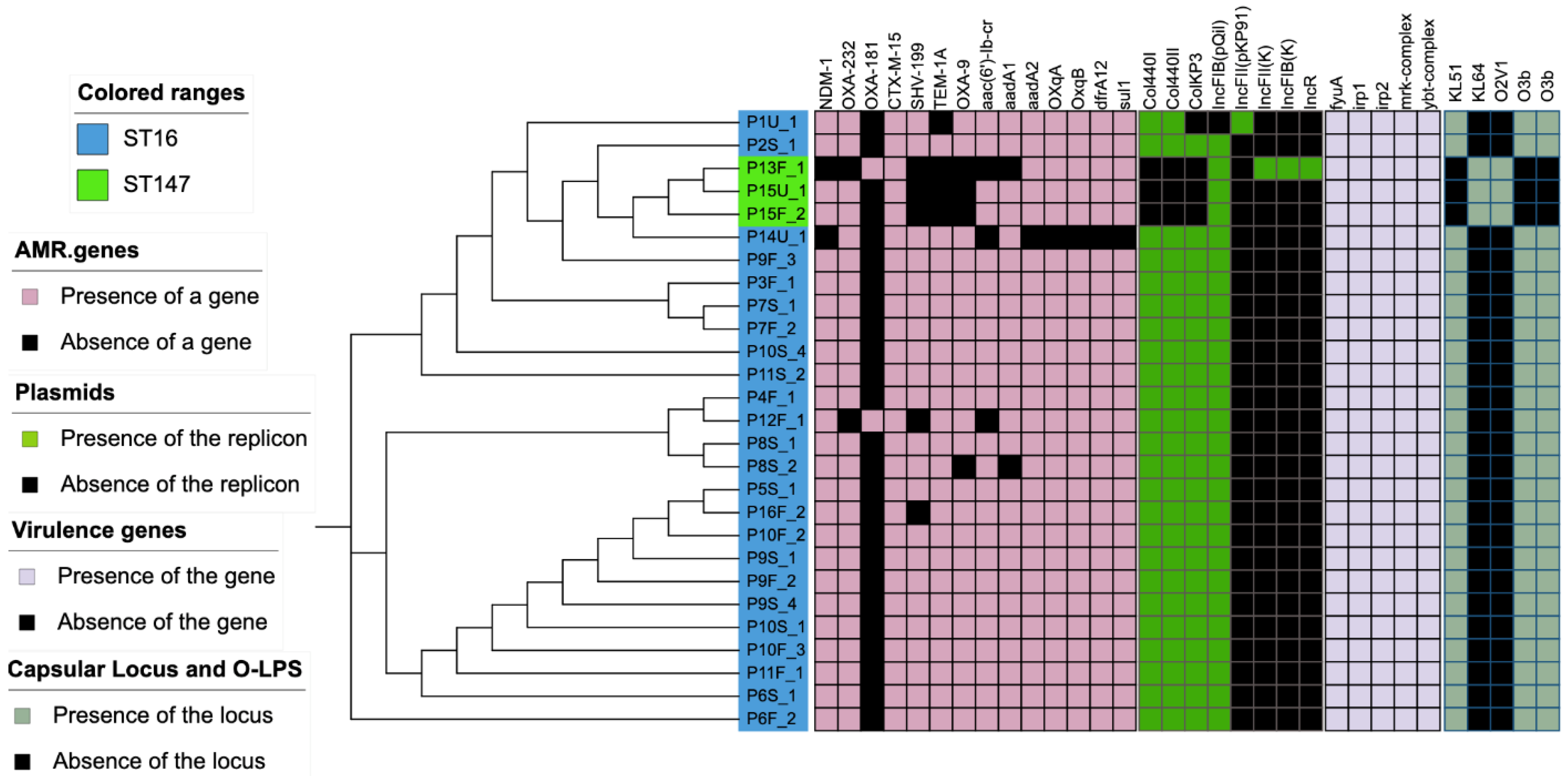
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lim, C.; Takahashi, E.; Hongsuwan, M.; Wuthiekanun, V.; Thamlikitkul, V.; Hinjoy, S.; Day, N.P.; Peacock, S.J.; Limmathurotsakul, D. Epidemiology and burden of multidrug-resistant bacterial infection in a developing country. eLife 2016, 5, e18082. [Google Scholar] [CrossRef]
- Suwantarat, N.; Carroll, K.C. Epidemiology and molecular characterization of multidrug-resistant Gram-negative bacteria in Southeast Asia. Antimicrob. Resist. Infect. Control 2016, 5, 15. [Google Scholar] [CrossRef] [Green Version]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Kattula, D.; Burkert, F. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery and Development of New Antibiotics; World Health Organization (WHO) Report: Geneva, Switzerland, 2017. [Google Scholar]
- Jitmuang, A.; Naksanguan, T.; Sirijatuphat, R.; Supapueng, O.; Kiratisin, P.; Thamlikitkul, V. Implementation of the World Health Organization’s Global Antimicrobial Resistance Surveillance System (GLASS) for the Surveillance of Sputum Specimens Collected from Patients at Siriraj Hospital. J. Med. Assoc. Thail. 2020, 103, 198–209. [Google Scholar]
- Sirijatuphat, R.; Pongsuttiyakorn, S.; Supapueng, O.; Kiratisin, P.; Thamlikitkul, V. Implementation of global antimicrobial resistance surveillance system (GLASS) in patients with bacteriuria. J. Glob. Antimicrob. Resist. 2020, 20, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Sirijatuphat, R.; Sripanidkulchai, K.; Boonyasiri, A.; Rattanaumpawan, P.; Supapueng, O.; Kiratisin, P.; Thamlikitkul, V. Implementation of global antimicrobial resistance surveillance system (GLASS) in patients with bacteremia. PLoS ONE 2018, 13, e0190132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aryal, P.U.; Thamjarungwong, B.; Singkhamanan, K.; Thongsuksai, P.; Ingviya, N.; Laohaprertthisan, V.; Darayon, R.; Yingkajorn, M. Emergence of Carbapenem-Resistant Enterobacteriaceae in a Tertiary Care Hospital in Southern Thailand†. Walailak J. Sci. Technol. 2020, 17, 1139–1148. [Google Scholar] [CrossRef]
- Chotiprasitsakul, D.; Srichatrapimuk, S.; Kirdlarp, S.; Pyden, A.D.; Santanirand, P. Epidemiology of carbapenem-resistant Enterobacteriaceae: A 5-year experience at a tertiary care hospital. Infect. Drug Resist. 2019, 12, 461–468. [Google Scholar] [CrossRef] [Green Version]
- Netikul, T.; Kiratisin, P. Genetic Characterization of Carbapenem-Resistant Enterobacteriaceae and the Spread of Carbapenem-Resistant Klebsiella pneumonia ST340 at a University Hospital in Thailand. PLoS ONE 2015, 10, e0139116. [Google Scholar] [CrossRef]
- Allegranzi, B.; Nejad, S.B.; Combescure, C.; Graafmans, W.; Attar, H.; Donaldson, L.; Pittet, D. Burden of endemic health-care-associated infection in developing countries: Systematic review and meta-analysis. Lancet 2011, 377, 228–241. [Google Scholar] [CrossRef]
- Boonyasiri, A.; Jauneikaite, E.; Brinkac, L.M.; Greco, C.; Lerdlamyong, K.; Tangkoskul, T.; Nguyen, K.; Thamlikitkul, V.; Fouts, D.E. Genomic and clinical characterisation of multidrug-resistant carbapenemase-producing ST231 and ST16 Klebsiella pneumoniae isolates colonising patients at Siriraj hospital, Bangkok, Thailand from 2015 to 2017. BMC Infect. Dis. 2021, 21, 142. [Google Scholar] [CrossRef]
- Paveenkittiporn, W.; Lyman, M.; Biedron, C.; Chea, N.; Bunthi, C.; Kolwaite, A.; Janejai, N. Molecular epidemiology of carbapenem-resistant Enterobacterales in Thailand, 2016–2018. Antimicrob. Resist. Infect. Control 2021, 10, 88. [Google Scholar] [CrossRef]
- Yong, D.; Toleman, M.A.; Giske, C.G.; Cho, H.S.; Sundman, K.; Lee, K.; Walsh, T.R. Characterization of a New Metallo- -Lactamase Gene, blaNDM-1, and a Novel Erythromycin Esterase Gene Carried on a Unique Genetic Structure in Klebsiella pneumoniae Sequence Type 14 from India. Antimicrob. Agents Chemother. 2009, 53, 5046–5054. [Google Scholar] [CrossRef] [Green Version]
- Müller, V.; Westerlund, F. Optical DNA Mapping in Nanofluidic Channels: Principles and Applications. Lab Chip 2017, 17, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, A.N.; Emilsson, G.; Nyberg, L.K.; Noble, C.; Stadler, L.S.; Fritzsche, J.; Moore, E.R.B.; Tegenfeldt, J.O.; Ambjornsson, T.; Westerlund, F. Competitive binding-based optical DNA mapping for fast identification of bacteria—multi-ligand transfer matrix theory and experimental applications on Escherichia coli. Nucleic Acids Res. 2014, 42, e118. [Google Scholar] [CrossRef] [Green Version]
- Nyberg, L.K.; Persson, F.; Berg, J.; Bergström, J.; Fransson, E.; Olsson, L.; Persson, M.; Stålnacke, A.; Wigenius, J.; Tegenfeldt, J.O.; et al. A single-step competitive binding assay for mapping of single DNA molecules. Biochem. Biophys. Res. Commun. 2012, 417, 404–408. [Google Scholar] [CrossRef]
- Müller, V.; Rajer, F.; Frykholm, K.; Nyberg, L.K.; Quaderi, S.; Fritzsche, J.; Kristiansson, E.; Ambjörnsson, T.; Sandegren, L.; Westerlund, F. Direct identification of antibiotic resistance genes on single plasmid molecules using CRISPR/Cas9 in combination with optical DNA mapping. Sci. Rep. 2016, 6, 37938. [Google Scholar] [CrossRef]
- Nyberg, L.K.; Quaderi, S.; Emilsson, G.; Karami, N.; Lagerstedt, E.; Müller, V.; Noble, C.; Hammarberg, S.; Nilsson, A.N.; Sjöberg, F.; et al. Rapid identification of intact bacterial resistance plasmids via optical mapping of single DNA molecules. Sci. Rep. 2016, 6, 30410. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.-L.; Sewunet, T.; Kk, S.; Giske, C.G.; Westerlund, F. Optical maps of plasmids as a proxy for clonal spread of MDR bacteria: A case study of an outbreak in a rural Ethiopian hospital. J. Antimicrob. Chemother. 2020, 75, 2804–2811. [Google Scholar] [CrossRef] [PubMed]
- Bikkarolla, S.K.; Nordberg, V.; Rajer, F.; Müller, V.; Kabir, M.H.; Kk, S.; Dvirnas, A.; Ambjörnsson, T.; Giske, C.G.; Navér, L.; et al. Optical DNA Mapping Combined with Cas9-Targeted Resistance Gene Identification for Rapid Tracking of Resistance Plasmids in a Neonatal Intensive Care Unit Outbreak. mBio 2019, 10, e00347-19. [Google Scholar] [CrossRef] [Green Version]
- Müller, V.; Karami, N.; Nyberg, L.K.; Pichler, C.; Torche Pedreschi, P.C.; Quaderi, S.; Fritzsche, J.; Ambjörnsson, T.; Åhrén, C.; Westerlund, F. Rapid Tracing of Resistance Plasmids in a Nosocomial Outbreak Using Optical DNA Mapping. ACS Infect. Dis. 2016, 2, 322–328. [Google Scholar] [CrossRef]
- Lindblom, A.; KK, S.; Müller, V.; Öz, R.; Sandström, H.; Åhrén, C.; Westerlund, F.; Karami, N. Interspecies Plasmid Transfer Appears Rare in Sequential Infections with Extended-Spectrum β-Lactamase (ESBL)-Producing Enterobacteriaceae; Elsevier: Amsterdam, The Netherlands, 2019; Volume 93, p. 6. [Google Scholar]
- Karami, N.; KK, S.; Yazdanshenas, S.; Lin, Y.-L.; Jaén-Luchoro, D.; Ekedahl, E.; Parameshwaran, S.; Lindblom, A.; Åhrén, C.; Westerlund, F. Identity of blaCTX-M Carrying Plasmids in Sequential ESBL-E. coli Isolates from Patients with Recurrent Urinary Tract Infections. Microorganisms 2021, 9, 1138. [Google Scholar] [CrossRef]
- Alizadehheidari, M.; Werner, E.; Noble, C.; Reiter-Schad, M.; Nyberg, L.K.; Fritzsche, J.; Mehlig, B.; Tegenfeldt, J.O.; Ambjörnsson, T.; Persson, F.; et al. Nanoconfined Circular and Linear DNA: Equilibrium Conformations and Unfolding Kinetics. Macromolecules 2015, 48, 871–878. [Google Scholar] [CrossRef] [Green Version]
- Dvirnas, A.; Pichler, C.; Stewart, C.L.; Quaderi, S.; Nyberg, L.K.; Müller, V.; Kumar Bikkarolla, S.; Kristiansson, E.; Sandegren, L.; Westerlund, F.; et al. Facilitated sequence assembly using densely labeled optical DNA barcodes: A combinatorial auction approach. PLoS ONE 2018, 13, e0193900. [Google Scholar] [CrossRef] [Green Version]
- Avolio, M.; Vignaroli, C.; Crapis, M.; Camporese, A. Co-production of NDM-1 and OXA-232 by ST16 Klebsiella pneumoniae, Italy, 2016. Future Microbiol. 2017, 12, 1119–1122. [Google Scholar] [CrossRef]
- Andrey, D.O.; Pereira Dantas, P.; Martins, W.B.S.; Marques De Carvalho, F.; Almeida, L.G.P.; Sands, K.; Portal, E.; Sauser, J.; Cayô, R.; Nicolas, M.F.; et al. An Emerging Clone, Klebsiellapneumoniae Carbapenemase 2–Producing K. pneumoniae Sequence Type 16, Associated With High Mortality Rates in a CC258-Endemic Setting. Clin. Infect. Dis. 2020, 71, e141–e150. [Google Scholar] [CrossRef]




| Isolate Code | Sample Type | MLST Type | Plasmid with blaNDM-1 Gene (Kb) | Plasmid with blaCTX-M-15 Gene (kb) | Plasmid with OXA-232 Gene (kb) |
|---|---|---|---|---|---|
| P1U_1 | Urine | 16 | 131 | ||
| P2S_1 | Sputum | 16 | 136 | ||
| P3F_1 | Feces | 16 | 125 | ||
| P4F_1 | Feces | 16 | 130 | 127 | 76 |
| P5S_1 | Sputum | 16 | 128 | 128 | 72 |
| P6S_1 | Sputum | 16 | 114 | ||
| P6F_2 | Feces | 16 | 121 | ||
| P7S_1 | Sputum | 16 | 130 | ||
| P7F_2 | Feces | 16 | 124 | ||
| P8S_1 | Sputum | 16 | 134 | ||
| P8S_2 | Sputum | 16 | 122 | ||
| P9S_1 | Sputum | 16 | 129 | ||
| P9F_2 | Feces | 16 | 128 | ||
| P9F_3 | Feces | 16 | 125 | ||
| P9S_4 | Sputum | 16 | 123 | ||
| P10S_1 | Sputum | 16 | 130 | ||
| P10F_2 | Feces | 16 | 126 | ||
| P10F_3 | Feces | 16 | 130 | ||
| P10S_4 | Sputum | 16 | 131 | ||
| P11F_1 | Feces | 16 | 138 | ||
| P11S_2 | Sputum | 16 | 132 | 128 | 77 |
| P12F_1 | Feces | 16 | 130 | ||
| P13F_1 | Feces | 147 | 55 | ||
| P14U_1 | Urine | 16 | 124 | ||
| P15U_1 | Urine | 147 | 52 | ||
| P15F_2 | Feces | 147 | 85 | ||
| P16F_1 | Feces | 16 | 122 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
KK, S.; Sewunet, T.; Wangchinda, W.; Tangkoskul, T.; Thamlikitkul, V.; Giske, C.G.; Westerlund, F. Optical DNA Mapping of Plasmids Reveals Clonal Spread of Carbapenem-Resistant Klebsiella pneumoniae in a Large Thai Hospital. Antibiotics 2021, 10, 1029. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10091029
KK S, Sewunet T, Wangchinda W, Tangkoskul T, Thamlikitkul V, Giske CG, Westerlund F. Optical DNA Mapping of Plasmids Reveals Clonal Spread of Carbapenem-Resistant Klebsiella pneumoniae in a Large Thai Hospital. Antibiotics. 2021; 10(9):1029. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10091029
Chicago/Turabian StyleKK, Sriram, Tsegaye Sewunet, Walaiporn Wangchinda, Teerawit Tangkoskul, Visanu Thamlikitkul, Christian G. Giske, and Fredrik Westerlund. 2021. "Optical DNA Mapping of Plasmids Reveals Clonal Spread of Carbapenem-Resistant Klebsiella pneumoniae in a Large Thai Hospital" Antibiotics 10, no. 9: 1029. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10091029






