A Cross-Metabolomic Approach Shows that Wheat Interferes with Fluorescent Pseudomonas Physiology through Its Root Metabolites
Abstract
:1. Introduction
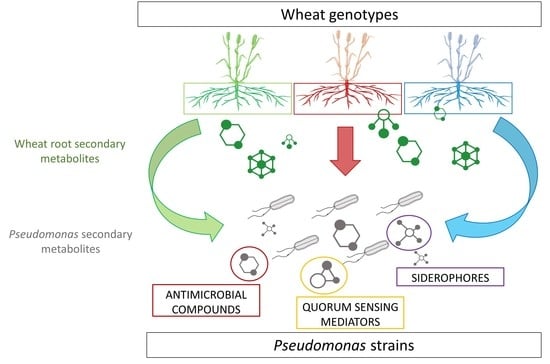
2. Results
2.1. Characterization of Root Extracts from Wheat Genotypes
2.2. Pseudomonas Secondary Metabolism Modifications in Response to Wheat Root Extracts
2.3. Focus on Secondary Metabolites Involved in Plant-Bacteria and Bacteria-Bacteria Interactions
3. Discussion
3.1. Wheat Root Metabolites Differ Depending on Plant Genotypes
3.2. Plant Root Compounds Affect the Biosynthesis of Bacterial Secondary Metabolites via a Signaling Effect
3.3. Wheat Root Extracts Interfere with the Production of Bacterial Secondary Metabolites Involved in Biotic Interactions
3.4. Wheat Root Extracts Modify the Synthesis of Siderophore and Compounds Whose Role in Plant-Bacteria Interactions Remains to Be Investigated
3.5. Despite Limitations, Our Approach Has Successfully Evidenced Key Metabolites Involved in Wheat-Pseudomonas Interactions
4. Materials and Methods
4.1. Bacterial Strains and Media
4.2. Extraction of Metabolites from Wheat Roots
4.3. Evaluation of the Effect of Wheat Root Extracts on Growth of Pseudomonas and on the Production of Bacterial Secondary Metabolites
4.4. LC-HRMS Analysis
4.5. Data Processing and Statistical Analysis of Secondary Metabolites of Wheat and Pseudomonas
4.6. Molecular Network Analysis and Identification of Metabolite
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Venturi, V.; Keel, C. Signaling in the rhizosphere. Trends Plant Sci. 2016, 21, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Bulgarelli, D.; Garrido-Oter, R.; Münch, P.C.; Weiman, A.; Dröge, J.; Pan, Y.; McHardy, A.C.; Schulze-Lefert, P. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 2015, 17, 392–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouffaud, M.-L.; Poirier, M.-A.; Muller, D.; Moënne-Loccoz, Y. Root microbiome relates to plant host evolution in maize and other Poaceae: Poaceae evolution and root bacteria. Environ. Microbiol. 2014, 16, 2804–2814. [Google Scholar] [CrossRef] [PubMed]
- Bais, H.P.; Weir, T.L.; Perry, L.G.; Gilroy, S.; Vivanco, J.M. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant. Biol. 2006, 57, 233–266. [Google Scholar] [CrossRef] [Green Version]
- Huang, A.C.; Jiang, T.; Liu, Y.-X.; Bai, Y.-C.; Reed, J.; Qu, B.; Goossens, A.; Nützmann, H.-W.; Bai, Y.; Osbourn, A. A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science 2019, 364. [Google Scholar] [CrossRef]
- Voges, M.J.E.E.E.; Bai, Y.; Schulze-Lefert, P.; Sattely, E.S. Plant-derived coumarins shape the composition of an Arabidopsis synthetic root microbiome. Proc. Natl. Acad. Sci. USA 2019, 116, 12558–12565. [Google Scholar] [CrossRef] [Green Version]
- Brink, S.C. Unlocking the secrets of the rhizosphere. Trends Plant Sci. 2016, 21, 169–170. [Google Scholar] [CrossRef] [Green Version]
- Stringlis, I.A.; Yu, K.; Feussner, K.; de Jonge, R.; van Bentum, S.; van Verk, M.C.; Berendsen, R.L.; Bakker, P.A.H.M.; Feussner, I.; Pieterse, C.M.J. MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc. Natl. Acad. Sci. USA 2018, 115, E5213–E5222. [Google Scholar] [CrossRef] [Green Version]
- Sasse, J.; Martinoia, E.; Northen, T. Feed your friends: Do plant exudates shape the root microbiome? Trends Plant Sci. 2018, 23, 25–41. [Google Scholar] [CrossRef] [Green Version]
- Cotton, T.E.A.; Pétriacq, P.; Cameron, D.D.; Meselmani, M.A.; Schwarzenbacher, R.; Rolfe, S.A.; Ton, J. Metabolic regulation of the maize rhizobiome by benzoxazinoids. ISME J. 2019, 13, 1647–1658. [Google Scholar] [CrossRef] [Green Version]
- Vacheron, J.; Desbrosses, G.; Bouffaud, M.-L.; Touraine, B.; Moënne-Loccoz, Y.; Muller, D.; Legendre, L.; Wisniewski-Dyé, F.; Prigent-Combaret, C. Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 2013, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weller, D.M. Pseudomonas biocontrol agents of soilborne pathogens: Looking back over 30 years. Phytopathology 2007, 97, 250–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almario, J.; Prigent-Combaret, C.; Muller, D.; Moënne-Loccoz, Y. Effect of clay mineralogy on iron bioavailability and rhizosphere transcription of 2,4-diacetylphloroglucinol biosynthetic genes in biocontrol Pseudomonas protegens. Mol. Plant Microbe Interact. 2013, 26, 566–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vacheron, J.; Moënne-Loccoz, Y.; Dubost, A.; Gonçalves-Martins, M.; Muller, D.; Prigent-Combaret, C. Fluorescent Pseudomonas strains with only few plant-beneficial properties are favored in the maize rhizosphere. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, L.; Robert, C.A.M.; Cadot, S.; Zhang, X.; Ye, M.; Li, B.; Manzo, D.; Chervet, N.; Steinger, T.; van der Heijden, M.G.A.; et al. Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 2018, 9, 2738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valente, J.; Gerin, F.; le Gouis, J.; Moënne-Loccoz, Y.; Prigent–Combaret, C. Ancient wheat varieties have a higher ability to interact with plant growth-promoting rhizobacteria. Plant Cell Environ. 2020, 43, 246–260. [Google Scholar] [CrossRef]
- Hay, A.-E.; Boubakri, H.; Buonomo, A.; Rey, M.; Meiffren, G.; Cotin-Galvan, L.; Comte, G.; Herrera-Belaroussi, A. Control of endophytic Frankia sporulation by Alnus nodule metabolites. Mol. Plant Microbe Interact. 2017, 30, 205–214. [Google Scholar] [CrossRef]
- Camilios-Neto, D.; Bonato, P.; Wassem, R.; Tadra-Sfeir, M.Z.; Brusamarello-Santos, L.C.; Valdameri, G.; Donatti, L.; Faoro, H.; Weiss, V.A.; Chubatsu, L.S.; et al. Dual RNA-seq transcriptional analysis of wheat roots colonized by Azospirillum brasilense reveals up-regulation of nutrient acquisition and cell cycle genes. BMC Genom. 2014, 15, 378. [Google Scholar] [CrossRef] [Green Version]
- Sessitsch, A.; Hardoim, P.; Döring, J.; Weilharter, A.; Krause, A.; Woyke, T.; Mitter, B.; Hauberg-Lotte, L.; Friedrich, F.; Rahalkar, M.; et al. Functional characteristics of an endophyte community colonizing rice roots as revealed by metagenomic analysis. Mol. Plant Microbe Interact. 2012, 25, 28–36. [Google Scholar] [CrossRef] [Green Version]
- Paungfoo-Lonhienne, C.; Lonhienne, T.G.; Yeoh, Y.K.; Donose, B.C.; Webb, R.I.; Parsons, J.; Liao, W.; Sagulenko, E.; Lakshmanan, P.; Hugenholtz, P.; et al. Crosstalk between sugarcane and a plant-growth promoting Burkholderia species. Sci. Rep. 2016, 6, 37389. [Google Scholar] [CrossRef]
- Balfourier, F.; Bouchet, S.; Robert, S.; de Oliveira, R.; Rimbert, H.; Kitt, J.; Choulet, F.; Paux, E. Worldwide phylogeography and history of wheat genetic diversity. Sci. Adv. 2019, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stutz, E.; Défago, G.; Kern, H. Naturally occurring fluorescent pseudomonads involved in suppression of black root rot of tobacco. Phytopathology 1986, 76, 181–185. [Google Scholar] [CrossRef]
- Shanahan, P.; O’Sullivan, D.J.; Simpson, P.; Glennon, J.D.; O’Gara, F. Isolation of 2,4-diacetylphloroglucinol from a fluorescent pseudomonad and investigation of physiological parameters influencing its production. Appl. Environ. Microbiol. 1992, 58, 353–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Besset-Manzoni, Y.; Rieusset, L.; Joly, P.; Comte, G.; Prigent-Combaret, C. Exploiting rhizosphere microbial cooperation for developing sustainable agriculture strategies. Environ. Sci. Pollut. Res. Int. 2018, 25, 29953–29970. [Google Scholar] [CrossRef] [PubMed]
- Gross, H.; Loper, J.E. Genomics of secondary metabolite production by Pseudomonas spp. Nat. Prod. Rep. 2009, 26, 1408. [Google Scholar] [CrossRef]
- Brazelton, J.N.; Pfeufer, E.E.; Sweat, T.A.; Gardener, B.B.M.; Coenen, C. 2,4-diacetylphloroglucinol alters plant root development. Mol. Plant Microbe Interact. 2008, 21, 1349–1358. [Google Scholar] [CrossRef] [Green Version]
- Spaepen, S.; Vanderleyden, J.; Remans, R. Indole-3-Acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 2007, 31, 425–448. [Google Scholar] [CrossRef] [Green Version]
- Rieusset, L.; Rey, M.; Muller, D.; Vacheron, J.; Gerin, F.; Dubost, A.; Comte, G.; Prigent-Combaret, C. Secondary metabolites from plant-associated Pseudomonas are overproduced in biofilm. Microb. Biotechnol. 2020, 13, 1562–1580. [Google Scholar] [CrossRef]
- Chin-A-Woeng, T.F.C.; Bloemberg, G.V.; Lugtenberg, B.J.J. Phenazines and their role in biocontrol by Pseudomonas bacteria. New Phytol. 2003, 157, 503–523. [Google Scholar] [CrossRef] [Green Version]
- Ye, L.; Cornelis, P.; Guillemyn, K.; Ballet, S.; Christophersen, C.; Hammerich, O. Structure revision of n -mercapto-4-formylcarbostyril produced by Pseudomonas fluorescens G308 to 2-(2-Hydroxyphenyl)Thiazole-4-Carbaldehyde [Aeruginaldehyde]. Nat. Prod. Commun. 2014, 9, 1934578X1400900. [Google Scholar] [CrossRef]
- Mark, G.L.; Dow, J.M.; Kiely, P.D.; Higgins, H.; Haynes, J.; Baysse, C.; Abbas, A.; Foley, T.; Franks, A.; Morrissey, J.; et al. Transcriptome profiling of bacterial responses to root exudates identifies genes involved in microbe-plant Interactions. Proc. Natl. Acad. Sci. USA 2005, 102, 17454–17459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, C.K.; Penesyan, A.; Hassan, K.A.; Loper, J.E.; Paulsen, I.T. Effect of tannic acid on the transcriptome of the soil bacterium Pseudomonas protegens Pf-5. Appl. Environ. Microbiol. 2013, 79, 3141–3145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Werra, P.; Huser, A.; Tabacchi, R.; Keel, C.; Maurhofer, M. Plant- and microbe-derived compounds affect the expression of genes encoding antifungal compounds in a pseudomonad with biocontrol activity. Appl. Environ. Microbiol. 2011, 77, 2807–2812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watrous, J.; Roach, P.; Alexandrov, T.; Heath, B.S.; Yang, J.Y.; Kersten, R.D.; van der Voort, M.; Pogliano, K.; Gross, H.; Raaijmakers, J.M.; et al. Mass spectral molecular networking of living microbial colonies. Proc. Natl. Acad. Sci. USA 2012, 109, E1743–E1752. [Google Scholar] [CrossRef] [Green Version]
- Olivon, F.; Elie, N.; Grelier, G.; Roussi, F.; Litaudon, M.; Touboul, D. Metgem software for the generation of molecular networks based on the t-sne algorithm. Anal. Chem. 2018, 90, 13900–13908. [Google Scholar] [CrossRef]
- Dinelli, G.; Segura-Carretero, A.; di Silvestro, R.; Marotti, I.; Arráez-Román, D.; Benedettelli, S.; Ghiselli, L.; Fernadez-Gutierrez, A. Profiles of phenolic compounds in modern and old common wheat varieties determined by liquid chromatography coupled with time-of-flight mass spectrometry. J. Chromatogr. A 2011, 1218, 7670–7681. [Google Scholar] [CrossRef]
- Meyer, T.; Renoud, S.; Vigouroux, A.; Miomandre, A.; Gaillard, V.; Kerzaon, I.; Prigent-Combaret, C.; Comte, G.; Moréra, S.; Vial, L.; et al. Regulation of hydroxycinnamic acid degradation drives Agrobacterium fabrum lifestyles. Mol. Plant Microbe Interact. 2018, 31, 814–822. [Google Scholar] [CrossRef] [Green Version]
- Macoy, D.M.; Kim, W.-Y.; Lee, S.Y.; Kim, M.G. Biosynthesis, physiology, and functions of hydroxycinnamic acid amides in plants. Plant Biotechnol. Rep. 2015, 9, 269–278. [Google Scholar] [CrossRef]
- Häusler, R.E.; Ludewig, F.; Krueger, S. Amino Acids—A life between metabolism and signaling. Plant Sci. 2014, 229, 225–237. [Google Scholar] [CrossRef]
- Valette, M.; Rey, M.; Gerin, F.; Comte, G.; Wisniewski-Dyé, F. A common metabolomic signature is observed upon inoculation of rice roots with various rhizobacteria. J. Integr. Plant Biol. 2020, 62, 228–246. [Google Scholar] [CrossRef]
- Wu, H.; Haig, T.; Pratley, J.; Lemerle, D.; An, M. Allelochemicals in wheat, (Triticum aestivum l.): Cultivar difference in the exudation of phenolic acids. J. Agric. Food Chem. 2001, 49, 3742–3745. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, A.; Sood, P.; Citovsky, V. The roles of plant phenolics in defence and communication during Agrobacterium and Rhizobium infection. Mol. Plant Pathol. 2010. [Google Scholar] [CrossRef]
- Chamam, A.; Wisniewski-Dyé, F.; Comte, G.; Bertrand, C.; Prigent-Combaret, C. Differential responses of Oryza Sativa secondary metabolism to biotic interactions with cooperative, commensal and phytopathogenic bacteria. Planta 2015, 242, 1439–1452. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Richter, A.; Jander, G. Beyond defense: Multiple functions of benzoxazinoids in maize metabolism. Plant Cell Physiol. 2018, 59, 1528–1533. [Google Scholar] [CrossRef] [PubMed]
- Marti, G.; Erb, M.; Boccard, J.; Glauser, G.; Doyen, G.R.; Villard, N.; Robert, C.A.M.; Turlings, T.C.J.; Rudaz, S.; Wolfender, J.-L. Metabolomics reveals herbivore-induced metabolites of resistance and susceptibility in maize leaves and roots: Plant-insect metabolomics. Plant Cell Environ. 2013, 36, 621–639. [Google Scholar] [CrossRef] [PubMed]
- Glauser, G.; Marti, G.; Villard, N.; Doyen, G.A.; Wolfender, J.-L.; Turlings, T.C.J.; Erb, M. Induction and detoxification of maize 1,4-benzoxazin-3-ones by insect herbivores: Defense induction and detoxification in maize. Plant J. 2011, 68, 901–911. [Google Scholar] [CrossRef]
- Niemeyer, H.M. Hydroxamic acids derived from 2-hydroxy-2 h-1,4-benzoxazin-3(4 h)-one: Key defense chemicals of cereals. J. Agric. Food Chem. 2009, 57, 1677–1696. [Google Scholar] [CrossRef]
- Wouters, F.C.; Gershenzon, J.; Vassão, D.G. Benzoxazinoids: Reactivity and modes of action of a versatile class of plant chemical defenses. J. Braz. Chem. Soc. 2016, 27, 1379–1397. [Google Scholar] [CrossRef]
- Neal, A.L.; Ahmad, S.; Gordon-Weeks, R.; Ton, J. Benzoxazinoids in root exudates of maize attract Pseudomonas putida to the rhizosphere. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [Green Version]
- Schütz, V.; Bigler, L.; Girel, S.; Laschke, L.; Sicker, D.; Schulz, M. Conversions of Benzoxazinoids and downstream metabolites by soil microorganisms. Front. Ecol. Evol. 2019, 7. [Google Scholar] [CrossRef] [Green Version]
- Schandry, N.; Becker, C. Allelopathic plants: Models for studying plant–interkingdom interactions. Trends Plant Sci. 2020, 25, 176–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelmohsen, U.R.; Grkovic, T.; Balasubramanian, S.; Kamel, M.S.; Quinn, R.J.; Hentschel, U. Elicitation of secondary metabolism in actinomycetes. Biotechnol. Adv. 2015, 33, 798–811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Redondo-Nieto, M.; Barret, M.; Morrissey, J.; Germaine, K.; Martínez-Granero, F.; Barahona, E.; Navazo, A.; Sánchez-Contreras, M.; Moynihan, J.A.; Muriel, C.; et al. Genome sequence reveals that Pseudomonas Fluorescens F113 possesses a large and diverse array of systems for rhizosphere function and host interaction. BMC Genom. 2013, 14, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mavrodi, D.V.; Blankenfeldt, W.; Thomashow, L.S. Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Annu. Rev. Phytopathol. 2006, 44, 417–445. [Google Scholar] [CrossRef] [PubMed]
- Jousset, A.; Rochat, L.; Lanoue, A.; Bonkowski, M.; Keel, C.; Scheu, S. Plants respond to pathogen infection by enhancing the antifungal gene expression of root-associated bacteria. Mol. Plant Microbe Interact. 2011, 24, 352–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rochat, L.; Péchy-Tarr, M.; Baehler, E.; Maurhofer, M.; Keel, C. Combination of fluorescent reporters for simultaneous monitoring of root colonization and antifungal gene expression by a biocontrol pseudomonad on cereals with flow cytometry. Mol. Plant Microbe Interact. 2010, 23, 949–961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haas, D.; Keel, C. Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu. Rev. Phytopathol. 2003, 41, 117–153. [Google Scholar] [CrossRef]
- Combes-Meynet, E.; Pothier, J.F.; Moënne-Loccoz, Y.; Prigent-Combaret, C. The Pseudomonas secondary metabolite 2,4-diacetylphloroglucinol is a signal inducing rhizoplane expression of Azospirillum genes involved in plant-growth promotion. Mol. Plant Microbe Interact. 2011, 24, 271–284. [Google Scholar] [CrossRef] [Green Version]
- Raaijmakers, J.M.; Mazzola, M. Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. Annu. Rev. Phytopathol. 2012, 50, 403–424. [Google Scholar] [CrossRef]
- Price-Whelan, A.; Dietrich, L.E.P.; Newman, D.K. Rethinking “secondary” metabolism: Physiological roles for phenazine antibiotics. Nat. Chem. Biol. 2006, 2, 71–78. [Google Scholar] [CrossRef]
- Mazzola, M.; Cook, R.J.; Thomashow, L.S.; Weller, D.M.; Pierson, L.S. Contribution of phenazine antibiotic biosynthesis to the ecological competence of fluorescent pseudomonads in soil habitats. Appl. Environ. Microbiol. 1992, 58, 2616–2624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maddula, V.S.R.K.; Pierson, E.A.; Pierson, L.S. Altering the ratio of phenazines in Pseudomonas chlororaphis, (aureofaciens) strain 30–84: Effects on biofilm formation and pathogen inhibition. J. Bacteriol. 2008, 190, 2759–2766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Yu, J.M.; Dorosky, R.J.; Pierson, L.S.; Pierson, E.A. The phenazine 2-hydroxy-phenazine-1-carboxylic acid promotes extracellular DNA release and has broad transcriptomic consequences in Pseudomonas chlororaphis 30–84. PLoS ONE 2016, 11, e0148003. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.M.; Wang, D.; Pierson, L.S.I.; Pierson, E.A. Effect of producing different phenazines on bacterial fitness and biological control in Pseudomonas chlororaphis 30–84. Plant Pathol. J. 2018, 34, 44–58. [Google Scholar] [CrossRef]
- Morohoshi, T.; Yamaguchi, T.; Xie, X.; Wang, W.; Takeuchi, K.; Someya, N. Complete genome sequence of Pseudomonas chlororaphis SubsP. aurantiaca reveals a triplicate quorum-sensing mechanism for regulation of phenazine production. Microbes Environ. 2017, 32, 47–53. [Google Scholar] [CrossRef] [Green Version]
- Keller, L.; Surette, M.G. Communication in bacteria: An ecological and evolutionary perspective. Nat. Rev. Microbiol. 2006, 4, 249–258. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Waters, C.M.; Bassler, B.L. Quorum sensing: Cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 2005, 21, 319–346. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, A.; Schmid, M.; van Tuinen, D.; Berg, G. Plant-driven selection of microbes. Plant Soil 2009, 321, 235–257. [Google Scholar] [CrossRef]
- Pérez-Montaño, F.; Guasch-Vidal, B.; González-Barroso, S.; López-Baena, F.J.; Cubo, T.; Ollero, F.J.; Gil-Serrano, A.M.; Rodríguez-Carvajal, M.Á.; Bellogín, R.A.; Espuny, M.R. Nodulation-gene-inducing flavonoids increase overall production of autoinducers and expression of n-acyl homoserine lactone synthesis genes in Rhizobia. Res. Microbiol. 2011, 162, 715–723. [Google Scholar] [CrossRef]
- Grandclément, C.; Tannières, M.; Moréra, S.; Dessaux, Y.; Faure, D. Quorum quenching: Role in nature and applied developments. FEMS Microbiol. Rev. 2016, 40, 86–116. [Google Scholar] [CrossRef] [PubMed]
- Elias, S.; Banin, E. Multi-species biofilms: Living with friendly neighbors. FEMS Microbiol. Rev. 2012, 36, 990–1004. [Google Scholar] [CrossRef] [PubMed]
- Selin, C.; Habibian, R.; Poritsanos, N.; Athukorala, S.N.P.; Fernando, D.; de Kievit, T.R. Phenazines are not essential for Pseudomonas chlororaphis pa23 biocontrol of Sclerotinia sclerotiorum, but do play a role in biofilm formation. FEMS Microbiol. Ecol. 2010, 71, 73–83. [Google Scholar] [CrossRef] [Green Version]
- González, J.F.; Venturi, V. A novel widespread interkingdom signaling circuit. Trends Plant Sci. 2013, 18, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Subramoni, S.; Gonzalez, J.F.; Johnson, A.; Péchy-Tarr, M.; Rochat, L.; Paulsen, I.; Loper, J.E.; Keel, C.; Venturi, V. Bacterial subfamily of luxr regulators that respond to plant compounds. Appl. Environ. Microbiol. 2011, 77, 4579–4588. [Google Scholar] [CrossRef] [Green Version]
- Youard, Z.A.; Mislin, G.L.A.; Majcherczyk, P.A.; Schalk, I.J.; Reimmann, C. Pseudomonas fluorescens CHA0 produces enantio-pyochelin, the optical antipode of the Pseudomonas aeruginosa siderophore pyochelin. J. Biol. Chem. 2007, 282, 35546–35553. [Google Scholar] [CrossRef] [Green Version]
- Audenaert, K.; Pattery, T.; Cornelis, P.; Höfte, M. Induction of systemic resistance to Botrytis cinerea in tomato by Pseudomonas aeruginosa 7nsk2: Role of salicylic acid, pyochelin, and pyocyanin. Mol. Plant Microbe Interact. 2002, 15, 1147–1156. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Wu, J.; Deng, Y.; Wang, J.; Wang, C.; Wang, J.; Chang, C.; Dong, Y.; Williams, P.; Zhang, L.-H. A Cell-cell communication signal integrates quorum sensing and stress response. Nat. Chem. Biol. 2013, 9, 339–343. [Google Scholar] [CrossRef]
- Cornelis, P. Putting an end to the Pseudomonas aeruginosa IQS controversy. Microbiol. Open 2020, 9. [Google Scholar] [CrossRef] [Green Version]
- Cortese, M.S.; Paszczynski, A.; Lewis, T.A.; Sebat, J.L.; Borek, V.; Crawford, R.L. Metal chelating properties of pyridine-2,6-bis(thiocarboxylic acid) produced by Pseudomonas spp. and the biological activities of the formed complexes. BioMetals 2002, 15, 103–120. [Google Scholar] [CrossRef]
- Budzikiewicz, H.; Lange, E.; Ockels, W. The mass spectral fragmentation behavior of pyridine carboxylic and thiocarboxylic acid esters. Phosphorus Sulfur Relat. Elem. 1981, 11, 33–45. [Google Scholar] [CrossRef]
- Budzikiewicz, H. Heteroaromatic monothiocarboxylic acids from Pseudomonas spp. Biodegradation 2003, 14, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Zawadzka, A.M.; Crawford, R.L.; Paszczynski, A.J. Pyridine-2,6-Bis(Thiocarboxylic Acid) produced by Pseudomonas stutzeri KC reduces and precipitates selenium and tellurium oxyanions. Appl. Environ. Microbiol. 2006, 72, 3119–3129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hildebrand, U.; Budzikiewicz, H. (+)-3,4-Dihydro-5-methyl-4-n-alkyl- und alkenyl-2H-pyrrole aus Pseudornonas putida. Z. Nat. 1986, 41, 1161–1169. [Google Scholar]
- Oburger, E.; Schmidt, H. New methods to unravel rhizosphere processes. Trends Plant Sci. 2016, 21, 243–255. [Google Scholar] [CrossRef] [PubMed]
- Walker, V.; Bertrand, C.; Bellvert, F.; Moënne-Loccoz, Y.; Bally, R.; Comte, G. Host plant secondary metabolite profiling shows a complex, strain-dependent response of maize to plant growth-promoting rhizobacteria of the genus Azospirillum. New Phytol. 2011, 189, 494–506. [Google Scholar] [CrossRef]
- Van Dam, N.M.; Bouwmeester, H.J. Metabolomics in the rhizosphere: Tapping into belowground chemical communication. Trends Plant Sci. 2016, 21, 256–265. [Google Scholar] [CrossRef]
- King, E.O.; Ward, M.K.; Raney, D.E. Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med. 1954, 44, 301–307. [Google Scholar]
- Bertani, G. Studies on lysogenesis I. J. Bacteriol. 1951, 62, 293–300. [Google Scholar] [CrossRef] [Green Version]
- El Zemrany, H.; Cortet, J.; Lutz, M.P.; Chabert, A.; Baudoin, E.; Haurat, J.; Maughan, N.; Félix, D.; Défago, G.; Bally, R.; et al. Field survival of the phytostimulator Azospirillum lipoferum CRT1 and functional impact on maize crop, biodegradation of crop residues, and soil faunal indicators in a context of decreasing nitrogen fertilisation. Soil Biol. Biochem. 2006, 38, 1712–1726. [Google Scholar] [CrossRef]
- Kessner, D.; Chambers, M.; Burke, R.; Agus, D.; Mallick, P. ProteoWizard: Open source software for rapid proteomics tools development. Bioinformatics 2008, 24, 2534–2536. [Google Scholar] [CrossRef]
- Giacomoni, F.; le Corguille, G.; Monsoor, M.; Landi, M.; Pericard, P.; Petera, M.; Duperier, C.; Tremblay-Franco, M.; Martin, J.-F.; Jacob, D.; et al. Workflow4Metabolomics: A collaborative research infrastructure for computational metabolomics. Bioinformatics 2015, 31, 1493–1495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Kuhl, C.; Tautenhahn, R.; Böttcher, C.; Larson, T.R.; Neumann, S. CAMERA: An integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry Data Sets. Anal. Chem. 2012, 84, 283–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thioulouse, J.; Chessel, D. ADE-4: Multivariate analysis and graphical display software. Stat. Comput. 1997, 7, 75–83. [Google Scholar] [CrossRef]
- Olivon, F.; Remy, S.; Grelier, G.; Apel, C.; Eydoux, C.; Guillemot, J.-C.; Neyts, J.; Delang, L.; Touboul, D.; Roussi, F.; et al. Antiviral compounds from Codiaeum peltatum targeted by a multi-informative molecular networks approach. J. Nat. Prod. 2019, 82, 330–340. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, C.; Zhao, X.; Xia, Y.; Sun, X.; Xie, W.; Xu, G. Deep annotation of hydroxycinnamic acid amides in plants based on ultra-high-performance liquid chromatography-high-resolution mass spectrometry and its in silico database. Anal. Chem. 2018, 90, 14321–14330. [Google Scholar] [CrossRef] [Green Version]
- Wolfender, J.-L.; Rudaz, S.; Choi, Y.H.; Kim, H.K. Plant metabolomics: From holistic data to relevant biomarkers. Curr. Med. Chem. 2013, 20, 1056–1090. [Google Scholar]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rieusset, L.; Rey, M.; Gerin, F.; Wisniewski-Dyé, F.; Prigent-Combaret, C.; Comte, G. A Cross-Metabolomic Approach Shows that Wheat Interferes with Fluorescent Pseudomonas Physiology through Its Root Metabolites. Metabolites 2021, 11, 84. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo11020084
Rieusset L, Rey M, Gerin F, Wisniewski-Dyé F, Prigent-Combaret C, Comte G. A Cross-Metabolomic Approach Shows that Wheat Interferes with Fluorescent Pseudomonas Physiology through Its Root Metabolites. Metabolites. 2021; 11(2):84. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo11020084
Chicago/Turabian StyleRieusset, Laura, Marjolaine Rey, Florence Gerin, Florence Wisniewski-Dyé, Claire Prigent-Combaret, and Gilles Comte. 2021. "A Cross-Metabolomic Approach Shows that Wheat Interferes with Fluorescent Pseudomonas Physiology through Its Root Metabolites" Metabolites 11, no. 2: 84. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo11020084