Combining Feature-Based Molecular Networking and Contextual Mass Spectral Libraries to Decipher Nutrimetabolomics Profiles
Abstract
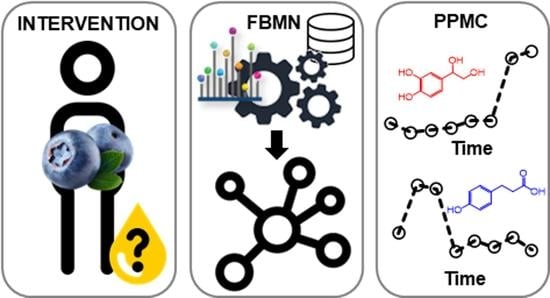
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Study Design, Sample Extraction, and LC-MS/MS Analysis
2.3. Data Pre-Processing
2.4. Data Availability: MassIVE Repository, Metadata and GNPS Jobs
- PI: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=a981ebd40809453ebe1524ff1fc8e265 (accessed on 27 June 2021).
- NI: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=0a239e71bb2045c292c4c96f4501249c (accessed on 27 June 2021).
2.5. “Nutri-Metabolomics” Library Building and Implementation
2.6. Molecular Networking Analyses
2.7. Analysis of Postprandial Kinetics
3. Results and Discussion
3.1. Optimization of the Input Parameters for Network Analysis
3.2. FBMN Annotation of NI and PI Datasets
3.3. VM and VC Relative Contributions to the Postprandial Metabolome
3.4. PPMC Analysis of Postprandial Kinetics
3.5. Nutrimetabolomics Outcomes from FBMN Molecular Networks
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted metabolomics strategies—Challenges and emerging directions. J. Am. Soc. Mass Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pomastowski, P.; Buszewski, B. Complementarity of matrix-and nanostructure-assisted laser desorption/ionization approaches. Nanomaterials 2019, 9, 260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaleckis, R.; Meister, I.; Zhang, P.; Wheelock, C.E. Challenges, progress and promises of metabolite annotation for LC–MS-based metabolomics. Curr. Opin. Biotechnol. 2019, 55, 44–50. [Google Scholar] [CrossRef]
- Kind, T.; Fiehn, O. Seven Golden Rules for heuristic filtering of molecular formulas obtained by accurate mass spectrometry. BMC Bioinform. 2007, 8, 105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [Green Version]
- Beniddir, M.A.; Kang, K.B.; Genta-Jouve, G.; Huber, F.; Rogers, S.; van der Hooft, J.J. Advances in decomposing complex metabolite mixtures using substructure-and network-based computational metabolomics approaches. Nat. Prod. Rep. 2021, 38, 1967–1993. [Google Scholar] [CrossRef] [PubMed]
- van der Hooft, J.J.; de Vos, R.C.; Mihaleva, V.; Bino, R.J.; Ridder, L.; de Roo, N.; Jacobs, D.M.; van Duynhoven, J.P.; Vervoort, J. Structural elucidation and quantification of phenolic conjugates present in human urine after tea intake. Anal. Chem. 2012, 84, 7263–7271. [Google Scholar] [CrossRef]
- Ramos, A.E.F.; Evanno, L.; Poupon, E.; Champy, P.; Beniddir, M.A. Natural products targeting strategies involving molecular networking: Different manners, one goal. Nat. Prod. Rep. 2019, 36, 960–980. [Google Scholar] [CrossRef]
- Aron, A.T.; Gentry, E.C.; McPhail, K.L.; Nothias, L.-F.; Nothias-Esposito, M.; Bouslimani, A.; Petras, D.; Gauglitz, J.M.; Sikora, N.; Vargas, F. Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nat. Protoc. 2020, 15, 1954–1991. [Google Scholar] [CrossRef]
- van der Hooft, J.J.; Padmanabhan, S.; Burgess, K.E.; Barrett, M.P. Urinary antihypertensive drug metabolite screening using molecular networking coupled to high-resolution mass spectrometry fragmentation. Metabolomics 2016, 12, 125. [Google Scholar] [CrossRef]
- Said, I.H.; Truex, J.D.; Heidorn, C.; Retta, M.B.; Petrov, D.D.; Haka, S.; Kuhnert, N. LC-MS/MS based molecular networking approach for the identification of cocoa phenolic metabolites in human urine. Food Res. Int. 2020, 132, 109119. [Google Scholar] [CrossRef] [PubMed]
- Nothias, L.-F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Oberleitner, D.; Schmid, R.; Schulz, W.; Bergmann, A.; Achten, C. Feature-based molecular networking for identification of organic micropollutants including metabolites by non-target analysis applied to riverbank filtration. Anal. Bioanal. Chem. 2021, 413, 5291–5300. [Google Scholar] [CrossRef]
- Padilla-González, G.F.; Sadgrove, N.J.; Ccana-Ccapatinta, G.V.; Leuner, O.; Fernandez-Cusimamani, E. Feature-based molecular networking to target the isolation of new caffeic acid esters from yacon (Smallanthus sonchifolius, Asteraceae). Metabolites 2020, 10, 407. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Mondragón, A.; Tuenter, E.; Ortiz, O.; Sakavitsi, M.E.; Nikou, T.; Halabalaki, M.; Caballero-George, C.; Apers, S.; Pieters, L.; Foubert, K. UPLC-MS/MS-based molecular networking and NMR structural determination for the untargeted phytochemical characterization of the fruit of Crescentia cujete (Bignoniaceae). Phytochemistry 2020, 177, 112438. [Google Scholar] [CrossRef]
- Xie, H.-F.; Kong, Y.-S.; Li, R.-Z.; Nothias, L.-F.; Melnik, A.V.; Zhang, H.; Liu, L.-L.; An, T.-T.; Liu, R.; Yang, Z. Feature-Based Molecular Networking Analysis of the Metabolites Produced by in vitro Solid-State Fermentation Reveals Pathways for the Bioconversion of Epigallocatechin Gallate. J. Agric. Food Chem. 2020, 68, 7995–8007. [Google Scholar] [CrossRef]
- Neto, F.C.; Raftery, D. Expanding Urinary Metabolite Annotation through Integrated Mass Spectral Similarity Networking. Anal. Chem. 2021, 93, 12001–12010. [Google Scholar] [CrossRef]
- Renai, L.; Ancillotti, C.; Ulaszewska, M.; Garcia-Aloy, M.; Mattivi, F.; Bartoletti, R.; Del Bubba, M. Comparison of chemometrics strategies for potential exposure markers discovery and false positive reduction in untargeted metabolomics: Application to the serum analysis by LC-HRMS after intake of Vaccinium fruits supplements. Anal. Bioanal. Chem. 2022, 414, 1841–1855. [Google Scholar] [CrossRef]
- Ancillotti, C.; Ulaszewska, M.; Mattivi, F.; Del Bubba, M. Untargeted metabolomics analytical strategy based on liquid chromatography/electrospray ionization linear ion trap quadrupole/orbitrap mass spectrometry for discovering new polyphenol metabolites in human biofluids after acute ingestion of vaccinium myrtillus berry supplement. J. Am. Soc. Mass Spectrom. 2019, 30, 381–402. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef]
- Vargas, F.; Weldon, K.C.; Sikora, N.; Wang, M.; Zhang, Z.; Gentry, E.C.; Panitchpakdi, M.W.; Caraballo-Rodríguez, A.M.; Dorrestein, P.C.; Jarmusch, A.K. Protocol for community-created public MS/MS reference spectra within the Global Natural Products Social Molecular Networking infrastructure. Rapid Commun. Mass Spectrom. 2020, 34, e8725. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Pellis, L.; van Erk, M.J.; van Ommen, B.; Bakker, G.C.; Hendriks, H.F.; Cnubben, N.H.; Kleemann, R.; van Someren, E.P.; Bobeldijk, I.; Rubingh, C.M. Plasma metabolomics and proteomics profiling after a postprandial challenge reveal subtle diet effects on human metabolic status. Metabolomics 2012, 8, 347–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Aloy, M.; Ulaszewska, M.; Franceschi, P.; Estruel-Amades, S.; Weinert, C.H.; Tor-Roca, A.; Urpi-Sarda, M.; Mattivi, F.; Andres-Lacueva, C. Discovery of intake biomarkers of lentils, chickpeas, and white beans by untargeted LC–MS metabolomics in serum and urine. Mol. Nutr. Food Res. 2020, 64, 1901137. [Google Scholar] [CrossRef]
- Iglesias-Carres, L.; Mas-Capdevila, A.; Bravo, F.I.; Aragonès, G.; Arola-Arnal, A.; Muguerza, B. A comparative study on the bioavailability of phenolic compounds from organic and nonorganic red grapes. Food Chem. 2019, 299, 125092. [Google Scholar] [CrossRef] [PubMed]
- Ancillotti, C.; Ciofi, L.; Rossini, D.; Chiuminatto, U.; Stahl-Zeng, J.; Orlandini, S.; Furlanetto, S.; Del Bubba, M. Liquid chromatographic/electrospray ionization quadrupole/time of flight tandem mass spectrometric study of polyphenolic composition of different Vaccinium berry species and their comparative evaluation. Anal. Bioanal. Chem. 2017, 409, 1347–1368. [Google Scholar] [CrossRef]
- de Mello, V.D.; Lankinen, M.A.; Lindström, J.; Puupponen-Pimiä, R.; Laaksonen, D.E.; Pihlajamäki, J.; Lehtonen, M.; Uusitupa, M.; Tuomilehto, J.; Kolehmainen, M. Fasting serum hippuric acid is elevated after bilberry (Vaccinium myrtillus) consumption and associates with improvement of fasting glucose levels and insulin secretion in persons at high risk of developing type 2 diabetes. Mol. Nutr. Food Res. 2017, 61, 1700019. [Google Scholar] [CrossRef]
- Vetrani, C.; Rivellese, A.A.; Annuzzi, G.; Adiels, M.; Borén, J.; Mattila, I.; Orešič, M.; Aura, A.-M. Metabolic transformations of dietary polyphenols: Comparison between in vitro colonic and hepatic models and in vivo urinary metabolites. J. Nutr. Biochem. 2016, 33, 111–118. [Google Scholar] [CrossRef]
- Aravind, S.M.; Wichienchot, S.; Tsao, R.; Ramakrishnan, S.; Chakkaravarthi, S. Role of dietary polyphenols on gut microbiota, their metabolites and health benefits. Food Res. Int. 2021, 142, 110189. [Google Scholar] [CrossRef]
- Stevens, J.F.; Maier, C.S. The chemistry of gut microbial metabolism of polyphenols. Phytochem. Rev. 2016, 15, 425–444. [Google Scholar] [CrossRef]
- Du, X.; Finn, C.E.; Qian, M.C. Volatile composition and odour-activity value of thornless ‘Black Diamond’ and ‘Marion’ blackberries. Food Chem. 2010, 119, 1127–1134. [Google Scholar] [CrossRef]
- Gutsche, B.; Grun, C.; Scheutzow, D.; Herderich, M. Tryptophan glycoconjugates in food and human urine. Biochem. J. 1999, 343, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Degu, A.; Ayenew, B.; Cramer, G.R.; Fait, A. Polyphenolic responses of grapevine berries to light, temperature, oxidative stress, abscisic acid and jasmonic acid show specific developmental-dependent degrees of metabolic resilience to perturbation. Food Chem. 2016, 212, 828–836. [Google Scholar] [CrossRef] [PubMed]
- Elsharif, S.A.; Buettner, A. Structure–odor relationship study on geraniol, nerol, and their synthesized oxygenated derivatives. J. Agric. Food Chem. 2016, 66, 2324–2333. [Google Scholar] [CrossRef] [PubMed]
- Colak, N.; Primetta, A.K.; Riihinen, K.R.; Jaakola, L.; Grúz, J.; Strnad, M.; Torun, H.; Ayaz, F.A. Phenolic compounds and antioxidant capacity in different-colored and non-pigmented berries of bilberry (Vaccinium myrtillus L.). Food Biosci. 2017, 20, 67–78. [Google Scholar] [CrossRef]
- Lotito, S.B.; Frei, B. Consumption of flavonoid-rich foods and increased plasma antioxidant capacity in humans: Cause, consequence, or epiphenomenon? Free Radic. Biol. Med. 2006, 41, 1727–1746. [Google Scholar] [CrossRef]
- Martínez-López, S.; Sarriá, B.; Gómez-Juaristi, M.; Goya, L.; Mateos, R.; Bravo-Clemente, L. Theobromine, caffeine, and theophylline metabolites in human plasma and urine after consumption of soluble cocoa products with different methylxanthine contents. Food Res. Int. 2014, 63, 446–455. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Croft, K. Vitamin E metabolism. Mol. Asp. Med. 2007, 28, 437–452. [Google Scholar] [CrossRef]




| Number of IDs | NI | PI |
|---|---|---|
| MZmine 1 | 26 | 49 |
| Statistical-based approach & manual annotation | 12 | 6 |
| FBMN | 24 | 43 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Renai, L.; Ulaszewska, M.; Mattivi, F.; Bartoletti, R.; Del Bubba, M.; van der Hooft, J.J.J. Combining Feature-Based Molecular Networking and Contextual Mass Spectral Libraries to Decipher Nutrimetabolomics Profiles. Metabolites 2022, 12, 1005. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo12101005
Renai L, Ulaszewska M, Mattivi F, Bartoletti R, Del Bubba M, van der Hooft JJJ. Combining Feature-Based Molecular Networking and Contextual Mass Spectral Libraries to Decipher Nutrimetabolomics Profiles. Metabolites. 2022; 12(10):1005. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo12101005
Chicago/Turabian StyleRenai, Lapo, Marynka Ulaszewska, Fulvio Mattivi, Riccardo Bartoletti, Massimo Del Bubba, and Justin J. J. van der Hooft. 2022. "Combining Feature-Based Molecular Networking and Contextual Mass Spectral Libraries to Decipher Nutrimetabolomics Profiles" Metabolites 12, no. 10: 1005. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo12101005