Co-Chaperones in Targeting and Delivery of Misfolded Proteins to the 26S Proteasome
Abstract
:1. Introduction
2. Protein Misfolding
3. Protein Quality Control
3.1. The Ubiquitin-Proteasome System (UPS)
3.2. Chaperones as Substrate Recognition Factors in Protein Quality Control
3.2.1. HSP70-Type Chaperones
3.2.2. HSP90-Type Chaperones
3.2.3. Chaperones in the Heat Shock Response
3.2.4. Molecular Chaperones and Protein Degradation
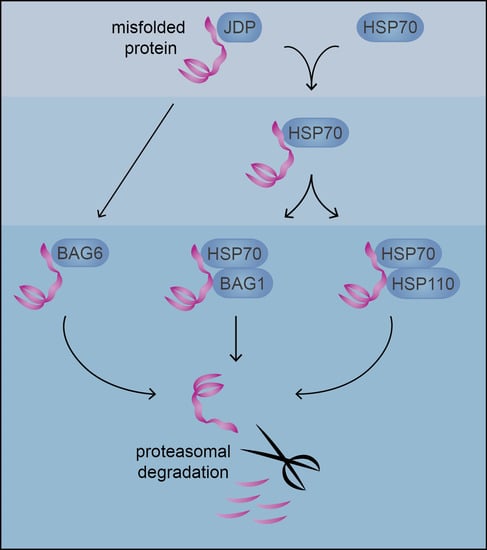
4. Co-Chaperones Decide the Fate of HSP70-Bound Substrates
4.1. Regulation of Chaperone Activity
4.2. Nucleotide Exchange Factors Direct Substrates to the Proteasome
4.2.1. Degradation through BAG-1
4.2.2. Degradation through BAG-6
4.2.3. Degradation through HSP110
4.3. Future Questions for Understanding the Role of Co-Chaperones as Shuttle-Factors for Misfolded Proteins
Funding
Acknowledgments
Conflicts of Interest
References
- Abildgaard, A.B.; Stein, A.; Nielsen, S.V.; Schultz-Knudsen, K.; Papaleo, E.; Shrikhande, A.; Hoffmann, E.R.; Bernstein, I.; Gerdes, A.-M.; Takahashi, M.; et al. Computational and cellular studies reveal structural destabilization and degradation of MLH1 variants in Lynch syndrome. eLife 2019, 8, e49138. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, S.V.; Stein, A.; Dinitzen, A.B.; Papaleo, E.; Tatham, M.H.; Poulsen, E.G.; Kassem, M.M.; Rasmussen, L.J.; Lindorff-Larsen, K.; Hartmann-Petersen, R. Predicting the impact of Lynch syndrome-causing missense mutations from structural calculations. PLoS Genet. 2017, 13, e1006739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheller, R.; Stein, A.; Nielsen, S.V.; Marin, F.I.; Gerdes, A.-M.; di Marco, M.; Papaleo, E.; Lindorff-Larsen, K.; Hartmann-Petersen, R. Toward mechanistic models for genotype-phenotype correlations in phenylketonuria using protein stability calculations. Hum. Mutat. 2019, 40, 444–457. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; van Dyk, D.; Li, Y.; Andrews, B.; Rao, H. A genome-wide synthetic dosage lethality screen reveals multiple pathways that require the functioning of ubiquitin-binding proteins Rad23 and Dsk2. BMC Biol. 2009, 7, 75. [Google Scholar] [CrossRef] [Green Version]
- Verma, R.; Oania, R.; Graumann, J.; Deshaies, R.J. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell 2004, 118, 99–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hipp, M.S.; Park, S.-H.; Hartl, F.U. Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 2014, 24, 506–514. [Google Scholar] [CrossRef]
- Hipp, M.S.; Kasturi, P.; Hartl, F.U. The proteostasis network and its decline in ageing. Nat. Rev. Mol. Cell Biol. 2019, 20, 421–435. [Google Scholar] [CrossRef]
- Mogk, A.; Bukau, B.; Kampinga, H.H. Cellular handling of protein aggregates by disaggregation machines. Mol. Cell 2018, 69, 214–226. [Google Scholar] [CrossRef]
- Balch, W.E.; Morimoto, R.I.; Dillin, A.; Kelly, J.W. Adapting proteostasis for disease intervention. Science 2008, 319, 916–919. [Google Scholar] [CrossRef] [Green Version]
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef]
- Sebastian, R.M.; Shoulders, M.D. Chemical biology framework to illuminate proteostasis. Annu. Rev. Biochem. 2020, 89, 529–555. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, R.L. Energetics of protein folding. J. Mol. Biol. 2007, 371, 283–301. [Google Scholar] [CrossRef] [PubMed]
- Dobson, C.M.; Šali, A.; Karplus, M. Protein Folding: A Perspective from theory and experiment. Angew. Chem. Int. Ed. Engl. 1998, 37, 868–893. [Google Scholar] [CrossRef]
- Kauzmann, W. Some factors in the interpretation of protein denaturation. Adv. Protein Chem. 1959, 14, 1–63. [Google Scholar] [PubMed]
- Hartl, F.U. Molecular chaperones in cellular protein folding. Nature 1996, 381, 571–579. [Google Scholar] [CrossRef]
- Rebeaud, M.E.; Mallik, S.; Goloubinoff, P.; Tawfik, D.S. On the evolution of chaperones and co-chaperones and the exponential expansion of proteome complexity. bioRxiv 2020. [Google Scholar] [CrossRef]
- Williams, P.D.; Pollock, D.D.; Goldstein, R.A. Functionality and the evolution of marginal stability in proteins: Inferences from lattice simulations. Evol. Bioinform. Online 2007, 2, 91–101. [Google Scholar] [CrossRef]
- Kim, Y.E.; Hipp, M.S.; Bracher, A.; Hayer-Hartl, M.; Hartl, F.U. Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 2013, 82, 323–355. [Google Scholar] [CrossRef]
- Meacham, G.C.; Patterson, C.; Zhang, W.; Younger, J.M.; Cyr, D.M. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat. Cell Biol. 2001, 3, 100–105. [Google Scholar] [CrossRef]
- Qu, B.H.; Strickland, E.H.; Thomas, P.J. Localization and suppression of a kinetic defect in cystic fibrosis transmembrane conductance regulator folding. J. Biol. Chem. 1997, 272, 15739–15744. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Venable, J.; LaPointe, P.; Hutt, D.M.; Koulov, A.V.; Coppinger, J.; Gurkan, C.; Kellner, W.; Matteson, J.; Plutner, H.; et al. Hsp90 Cochaperone Aha1 Downregulation Rescues Misfolding of CFTR in Cystic Fibrosis. Cell 2006, 127, 803–815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almstedt, K.; Lundqvist, M.; Carlsson, J.; Karlsson, M.; Persson, B.; Jonsson, B.-H.; Carlsson, U.; Hammarström, P. Unfolding a folding disease: Folding, misfolding and aggregation of the marble brain syndrome-associated mutant H107Y of human carbonic anhydrase II. J. Mol. Biol. 2004, 342, 619–633. [Google Scholar] [CrossRef] [PubMed]
- Arlow, T.; Scott, K.; Wagenseller, A.; Gammie, A. Proteasome inhibition rescues clinically significant unstable variants of the mismatch repair protein Msh2. Proc. Natl. Acad. Sci. USA 2013, 110, 246–251. [Google Scholar] [CrossRef] [Green Version]
- Hidvegi, T.; Schmidt, B.Z.; Hale, P.; Perlmutter, D.H. Accumulation of mutant alpha1-antitrypsin Z in the endoplasmic reticulum activates caspases-4 and -12, NFkappaB, and BAP31 but not the unfolded protein response. J. Biol. Chem. 2005, 280, 39002–39015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lomas, D.A.; Evans, D.L.; Finch, J.T.; Carrell, R.W. The mechanism of Z alpha 1-antitrypsin accumulation in the liver. Nature 1992, 357, 605–607. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Whitman, I.; Molmenti, E.; Moore, K.; Hippenmeyer, P.; Perlmutter, D.H. A lag in intracellular degradation of mutant alpha 1-antitrypsin correlates with the liver disease phenotype in homozygous PiZZ alpha 1-antitrypsin deficiency. Proc. Natl. Acad. Sci. USA 1994, 91, 9014–9018. [Google Scholar] [CrossRef] [Green Version]
- Chiti, F.; Dobson, C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006, 75, 333–366. [Google Scholar] [CrossRef] [Green Version]
- Lázaro, D.F.; Bellucci, A.; Brundin, P.; Outeiro, T.F. Editorial: Protein misfolding and spreading pathology in neurodegenerative diseases. Front. Mol. Neurosci. 2019, 12, 312. [Google Scholar] [CrossRef] [Green Version]
- Ross, C.A.; Poirier, M.A. Protein aggregation and neurodegenerative disease. Nat. Med. 2004, 10, S10–S17. [Google Scholar] [CrossRef]
- Soto, C. Unfolding the role of protein misfolding in neurodegenerative diseases. Nat. Rev. Neurosci. 2003, 4, 49–60. [Google Scholar] [CrossRef]
- Schneider, K.; Bertolotti, A. Surviving protein quality control catastrophes--from cells to organisms. J. Cell. Sci. 2015, 128, 3861–3869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rock, K.L.; Gramm, C.; Rothstein, L.; Clark, K.; Stein, R.; Dick, L.; Hwang, D.; Goldberg, A.L. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 1994, 78, 761–771. [Google Scholar] [CrossRef]
- Hershko, A.; Eytan, E.; Ciechanover, A.; Haas, A.L. Immunochemical analysis of the turnover of ubiquitin-protein conjugates in intact cells. Relationship to the breakdown of abnormal proteins. J. Biol. Chem. 1982, 257, 13964–13970. [Google Scholar]
- Ciechanover, A.; Finley, D.; Varshavsky, A. Ubiquitin dependence of selective protein degradation demonstrated in the mammalian cell cycle mutant ts85. Cell 1984, 37, 57–66. [Google Scholar] [CrossRef]
- Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009, 78, 477–513. [Google Scholar] [CrossRef] [Green Version]
- Schubert, U.; Antón, L.C.; Gibbs, J.; Norbury, C.C.; Yewdell, J.W.; Bennink, J.R. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 2000, 404, 770–774. [Google Scholar] [CrossRef]
- Ciechanover, A.; Heller, H.; Elias, S.; Haas, A.L.; Hershko, A. ATP-dependent conjugation of reticulocyte proteins with the polypeptide required for protein degradation. Proc. Natl. Acad. Sci. USA 1980, 77, 1365–1368. [Google Scholar] [CrossRef] [Green Version]
- Ciechanover, A.; Elias, S.; Heller, H.; Ferber, S.; Hershko, A. Characterization of the heat-stable polypeptide of the ATP-dependent proteolytic system from reticulocytes. J. Biol. Chem. 1980, 255, 7525–7528. [Google Scholar]
- Ciehanover, A.; Hod, Y.; Hershko, A. A heat-stable polypeptide component of an ATP-dependent proteolytic system from reticulocytes. Biochem. Biophys. Res. Commun. 1978, 81, 1100–1105. [Google Scholar] [CrossRef]
- Wilkinson, K.D.; Urban, M.K.; Haas, A.L. Ubiquitin is the ATP-dependent proteolysis factor I of rabbit reticulocytes. J. Biol. Chem. 1980, 255, 7529–7532. [Google Scholar]
- Hershko, A.; Ciechanover, A.; Rose, I.A. Identification of the active amino acid residue of the polypeptide of ATP-dependent protein breakdown. J. Biol. Chem. 1981, 256, 1525–1528. [Google Scholar] [PubMed]
- Chau, V.; Tobias, J.W.; Bachmair, A.; Marriott, D.; Ecker, D.J.; Gonda, D.K.; Varshavsky, A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 1989, 243, 1576–1583. [Google Scholar] [CrossRef] [PubMed]
- Finley, D.; Sadis, S.; Monia, B.P.; Boucher, P.; Ecker, D.J.; Crooke, S.T.; Chau, V. Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol. Cell. Biol. 1994, 14, 5501–5509. [Google Scholar] [CrossRef] [PubMed]
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef]
- Hershko, A.; Ciechanover, A.; Heller, H.; Haas, A.L.; Rose, I.A. Proposed role of ATP in protein breakdown: Conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis. Proc. Natl. Acad. Sci. USA 1980, 77, 1783–1786. [Google Scholar] [CrossRef] [Green Version]
- Thrower, J.S.; Hoffman, L.; Rechsteiner, M.; Pickart, C.M. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000, 19, 94–102. [Google Scholar] [CrossRef] [Green Version]
- Hershko, A.; Heller, H.; Elias, S.; Ciechanover, A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J. Biol. Chem. 1983, 258, 8206–8214. [Google Scholar]
- Li, W.; Bengtson, M.H.; Ulbrich, A.; Matsuda, A.; Reddy, V.A.; Orth, A.; Chanda, S.K.; Batalov, S.; Joazeiro, C.A.P. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS ONE 2008, 3, e1487. [Google Scholar] [CrossRef]
- Eisele, F.; Wolf, D.H. Degradation of misfolded protein in the cytoplasm is mediated by the ubiquitin ligase Ubr1. FEBS Lett. 2008, 582, 4143–4146. [Google Scholar] [CrossRef] [Green Version]
- Gardner, R.G.; Nelson, Z.W.; Gottschling, D.E. Degradation-mediated protein quality control in the nucleus. Cell 2005, 120, 803–815. [Google Scholar] [CrossRef] [Green Version]
- Heck, J.W.; Cheung, S.K.; Hampton, R.Y. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc. Natl. Acad. Sci. USA 2010, 107, 1106–1111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Connell, P.; Ballinger, C.A.; Jiang, J.; Wu, Y.; Thompson, L.J.; Höhfeld, J.; Patterson, C. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 2001, 3, 93–96. [Google Scholar] [CrossRef] [PubMed]
- Demand, J.; Alberti, S.; Patterson, C.; Höhfeld, J. Cooperation of a ubiquitin domain protein and an E3 ubiquitin ligase during chaperone/proteasome coupling. Curr. Biol. 2001, 11, 1569–1577. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.; Ballinger, C.A.; Wu, Y.; Dai, Q.; Cyr, D.M.; Höhfeld, J.; Patterson, C. CHIP is a U-box-dependent E3 ubiquitin ligase: Identification of Hsc70 as a target for ubiquitylation. J. Biol. Chem. 2001, 276, 42938–42944. [Google Scholar] [CrossRef] [Green Version]
- Bard, J.A.M.; Goodall, E.A.; Greene, E.R.; Jonsson, E.; Dong, K.C.; Martin, A. Structure and function of the 26S proteasome. Annu. Rev. Biochem. 2018, 87, 697–724. [Google Scholar] [CrossRef]
- Collins, G.A.; Goldberg, A.L. The logic of the 26S proteasome. Cell 2017, 169, 792–806. [Google Scholar] [CrossRef] [Green Version]
- Glickman, M.H.; Rubin, D.M.; Coux, O.; Wefes, I.; Pfeifer, G.; Cjeka, Z.; Baumeister, W.; Fried, V.A.; Finley, D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 1998, 94, 615–623. [Google Scholar] [CrossRef] [Green Version]
- Deveraux, Q.; Ustrell, V.; Pickart, C.; Rechsteiner, M. A 26 S protease subunit that binds ubiquitin conjugates. J. Biol. Chem. 1994, 269, 7059–7061. [Google Scholar]
- Husnjak, K.; Elsasser, S.; Zhang, N.; Chen, X.; Randles, L.; Shi, Y.; Hofmann, K.; Walters, K.J.; Finley, D.; Dikic, I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 2008, 453, 481–488. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Chen, X.; Elsasser, S.; Stocks, B.B.; Tian, G.; Lee, B.-H.; Shi, Y.; Zhang, N.; de Poot, S.A.H.; Tuebing, F.; et al. Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science 2016, 351, aad9421. [Google Scholar] [CrossRef] [Green Version]
- Elsasser, S.; Gali, R.R.; Schwickart, M.; Larsen, C.N.; Leggett, D.S.; Müller, B.; Feng, M.T.; Tübing, F.; Dittmar, G.A.G.; Finley, D. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat. Cell Biol. 2002, 4, 725–730. [Google Scholar] [CrossRef] [PubMed]
- Funakoshi, M.; Sasaki, T.; Nishimoto, T.; Kobayashi, H. Budding yeast Dsk2p is a polyubiquitin-binding protein that can interact with the proteasome. Proc. Natl. Acad. Sci. USA 2002, 99, 745–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madsen, L.; Schulze, A.; Seeger, M.; Hartmann-Petersen, R. Ubiquitin domain proteins in disease. BMC Biochem. 2007, 8, S1. [Google Scholar] [CrossRef] [Green Version]
- Rosenzweig, R.; Bronner, V.; Zhang, D.; Fushman, D.; Glickman, M.H. Rpn1 and Rpn2 coordinate ubiquitin processing factors at proteasome. J. Biol. Chem. 2012, 287, 14659–14671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schauber, C.; Chen, L.; Tongaonkar, P.; Vega, I.; Lambertson, D.; Potts, W.; Madura, K. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature 1998, 391, 715–718. [Google Scholar] [CrossRef] [PubMed]
- Andersen, K.M.; Hofmann, K.; Hartmann-Petersen, R. Ubiquitin-binding proteins: Similar, but different. Essays Biochem. 2005, 41, 49–67. [Google Scholar] [CrossRef] [Green Version]
- Hofmann, K.; Bucher, P. The UBA domain: A sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem. Sci. 1996, 21, 172–173. [Google Scholar] [CrossRef]
- Wilkinson, C.R.M.; Seeger, M.; Hartmann-Petersen, R.; Stone, M.; Wallace, M.; Semple, C.; Gordon, C. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat. Cell Biol. 2001, 3, 939–943. [Google Scholar] [CrossRef]
- Verma, R.; Aravind, L.; Oania, R.; McDonald, W.H.; Yates, J.R.; Koonin, E.V.; Deshaies, R.J. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 2002, 298, 611–615. [Google Scholar] [CrossRef]
- Yao, T.; Cohen, R.E. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 2002, 419, 403–407. [Google Scholar] [CrossRef]
- Park, K.C.; Woo, S.K.; Yoo, Y.J.; Wyndham, A.M.; Baker, R.T.; Chung, C.H. Purification and characterization of UBP6, a new ubiquitin-specific protease in saccharomyces cerevisiae. Arch. Biochem. Biophys. 1997, 347, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Naqvi, N.I.; Yang, H.; Teo, T.S. Identification of a 26S proteasome-associated UCH in fission yeast. Biochem. Biophys. Res. Commun. 2000, 272, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Lee, B.; King, R.W.; Finley, D.; Kirschner, M.W. Substrate degradation by the proteasome: A single-molecule kinetic analysis. Science 2015, 348, 1250834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braun, B.C.; Glickman, M.; Kraft, R.; Dahlmann, B.; Kloetzel, P.M.; Finley, D.; Schmidt, M. The base of the proteasome regulatory particle exhibits chaperone-like activity. Nat. Cell Biol. 1999, 1, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Groll, M.; Ditzel, L.; Löwe, J.; Stock, D.; Bochtler, M.; Bartunik, H.D.; Huber, R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature 1997, 386, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Groll, M.; Bajorek, M.; Köhler, A.; Moroder, L.; Rubin, D.M.; Huber, R.; Glickman, M.H.; Finley, D. A gated channel into the proteasome core particle. Nat. Struct. Biol. 2000, 7, 1062–1067. [Google Scholar] [CrossRef]
- Smith, D.M.; Chang, S.-C.; Park, S.; Finley, D.; Cheng, Y.; Goldberg, A.L. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol. Cell 2007, 27, 731–744. [Google Scholar] [CrossRef] [Green Version]
- Finley, D.; Chen, X.; Walters, K.J. Gates, channels, and switches: Elements of the proteasome machine. Trends Biochem. Sci. 2016, 41, 77–93. [Google Scholar] [CrossRef] [Green Version]
- Kisselev, A.F.; Akopian, T.N.; Woo, K.M.; Goldberg, A.L. The sizes of peptides generated from protein by mammalian 26 and 20 S proteasomes. Implications for understanding the degradative mechanism and antigen presentation. J. Biol. Chem. 1999, 274, 3363–3371. [Google Scholar] [CrossRef] [Green Version]
- Michalek, M.T.; Grant, E.P.; Gramm, C.; Goldberg, A.L.; Rock, K.L. A role for the ubiquitin-dependent proteolytic pathway in MHC class I-restricted antigen presentation. Nature 1993, 363, 552–554. [Google Scholar] [CrossRef]
- Comyn, S.A.; Young, B.P.; Loewen, C.J.; Mayor, T. Prefoldin promotes proteasomal degradation of cytosolic proteins with missense mutations by maintaining substrate solubility. PLoS Genet. 2016, 12, e1006184. [Google Scholar] [CrossRef] [PubMed]
- Stolz, A.; Hilt, W.; Buchberger, A.; Wolf, D.H. Cdc48: A power machine in protein degradation. Trends Biochem. Sci. 2011, 36, 515–523. [Google Scholar] [CrossRef] [PubMed]
- Fourie, A.M.; Sambrook, J.F.; Gething, M.J. Common and divergent peptide binding specificities of hsp70 molecular chaperones. J. Biol. Chem. 1994, 269, 30470–30478. [Google Scholar] [PubMed]
- Gragerov, A.; Gottesman, M.E. Different peptide binding specificities of hsp70 family members. J. Mol. Biol. 1994, 241, 133–135. [Google Scholar] [CrossRef] [PubMed]
- Kampinga, H.H.; Craig, E.A. The Hsp70 chaperone machinery: J-proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 2010, 11, 579–592. [Google Scholar] [CrossRef] [Green Version]
- Sharma, D.; Masison, D.C. Single methyl group determines prion propagation and protein degradation activities of yeast heat shock protein (Hsp)-70 chaperones Ssa1p and Ssa2p. Proc. Natl. Acad. Sci. USA 2011, 108, 13665–13670. [Google Scholar] [CrossRef] [Green Version]
- Rosenzweig, R.; Nillegoda, N.B.; Mayer, M.P.; Bukau, B. The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 2019, 20, 665–680. [Google Scholar] [CrossRef]
- Ryu, S.W.; Stewart, R.; Pectol, D.C.; Ender, N.A.; Wimalarathne, O.; Lee, J.-H.; Zanini, C.P.; Harvey, A.; Huibregtse, J.M.; Mueller, P.; et al. Proteome-wide identification of HSP70/HSC70 chaperone clients in human cells. PLoS Biol. 2020, 18, e3000606. [Google Scholar] [CrossRef]
- Chappell, T.G.; Konforti, B.B.; Schmid, S.L.; Rothman, J.E. The ATPase core of a clathrin uncoating protein. J. Biol. Chem. 1987, 262, 746–751. [Google Scholar]
- Wang, T.F.; Chang, J.H.; Wang, C. Identification of the peptide binding domain of hsc70. 18-Kilodalton fragment located immediately after ATPase domain is sufficient for high affinity binding. J. Biol. Chem. 1993, 268, 26049–26051. [Google Scholar]
- Zhu, X.; Zhao, X.; Burkholder, W.F.; Gragerov, A.; Ogata, C.M.; Gottesman, M.E.; Hendrickson, W.A. Structural analysis of substrate binding by the molecular chaperone DnaK. Science 1996, 272, 1606–1614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swain, J.F.; Dinler, G.; Sivendran, R.; Montgomery, D.L.; Stotz, M.; Gierasch, L.M. Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol. Cell 2007, 26, 27–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flaherty, K.M.; DeLuca-Flaherty, C.; McKay, D.B. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature 1990, 346, 623–628. [Google Scholar] [CrossRef]
- McCarty, J.S.; Buchberger, A.; Reinstein, J.; Bukau, B. The role of ATP in the functional cycle of the DnaK chaperone system. J. Mol. Biol. 1995, 249, 126–137. [Google Scholar] [CrossRef] [PubMed]
- Buchberger, A.; Theyssen, H.; Schröder, H.; McCarty, J.S.; Virgallita, G.; Milkereit, P.; Reinstein, J.; Bukau, B. Nucleotide-induced conformational changes in the ATPase and substrate binding domains of the DnaK chaperone provide evidence for interdomain communication. J. Biol. Chem. 1995, 270, 16903–16910. [Google Scholar] [CrossRef] [Green Version]
- Qi, R.; Sarbeng, E.B.; Liu, Q.; Le, K.Q.; Xu, X.; Xu, H.; Yang, J.; Wong, J.L.; Vorvis, C.; Hendrickson, W.A.; et al. Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP. Nat. Struct. Mol. Biol. 2013, 20, 900–907. [Google Scholar] [CrossRef] [Green Version]
- Szabo, A.; Langer, T.; Schröder, H.; Flanagan, J.; Bukau, B.; Hartl, F.U. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK, DnaJ, and GrpE. Proc. Natl. Acad. Sci. USA 1994, 91, 10345–10349. [Google Scholar] [CrossRef] [Green Version]
- Rüdiger, S.; Germeroth, L.; Schneider-Mergener, J.; Bukau, B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 1997, 16, 1501–1507. [Google Scholar] [CrossRef] [Green Version]
- Flynn, G.C.; Pohl, J.; Flocco, M.T.; Rothman, J.E. Peptide-binding specificity of the molecular chaperone BiP. Nature 1991, 353, 726–730. [Google Scholar] [CrossRef]
- Gragerov, A.; Zeng, L.; Zhao, X.; Burkholder, W.; Gottesman, M.E. Specificity of DnaK-peptide binding. J. Mol. Biol. 1994, 235, 848–854. [Google Scholar] [CrossRef]
- Houben, B.; Michiels, E.; Ramakers, M.; Konstantoulea, K.; Louros, N.; Verniers, J.; van der Kant, R.; de Vleeschouwer, M.; Chicória, N.; Vanpoucke, T.; et al. Autonomous aggregation suppression by acidic residues explains why chaperones favour basic residues. EMBO J. 2020, 39, e102864. [Google Scholar] [CrossRef]
- Schlecht, R.; Erbse, A.H.; Bukau, B.; Mayer, M.P. Mechanics of Hsp70 chaperones enables differential interaction with client proteins. Nat. Struct. Mol. Biol. 2011, 18, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Kellner, R.; Hofmann, H.; Barducci, A.; Wunderlich, B.; Nettels, D.; Schuler, B. Single-molecule spectroscopy reveals chaperone-mediated expansion of substrate protein. Proc. Natl. Acad. Sci. USA 2014, 111, 13355–13360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palleros, D.R.; Shi, L.; Reid, K.L.; Fink, A.L. hsp70-protein complexes. Complex stability and conformation of bound substrate protein. J. Biol. Chem. 1994, 269, 13107–13114. [Google Scholar] [PubMed]
- Sekhar, A.; Rosenzweig, R.; Bouvignies, G.; Kay, L.E. Mapping the conformation of a client protein through the Hsp70 functional cycle. Proc. Natl. Acad. Sci. USA 2015, 112, 10395–10400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mashaghi, A.; Bezrukavnikov, S.; Minde, D.P.; Wentink, A.S.; Kityk, R.; Zachmann-Brand, B.; Mayer, M.P.; Kramer, G.; Bukau, B.; Tans, S.J. Alternative modes of client binding enable functional plasticity of Hsp70. Nature 2016, 539, 448–451. [Google Scholar] [CrossRef]
- Sekhar, A.; Rosenzweig, R.; Bouvignies, G.; Kay, L.E. Hsp70 biases the folding pathways of client proteins. Proc. Natl. Acad. Sci. USA 2016, 113, E2794–E2801. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Zhang, D.; Hughes, C.; Okuno, Y.; Sekhar, A.; Cavagnero, S. Heterogeneous binding of the SH3 client protein to the DnaK molecular chaperone. Proc. Natl. Acad. Sci. USA 2015, 112, E4206–E4215. [Google Scholar] [CrossRef] [Green Version]
- Morán Luengo, T.; Kityk, R.; Mayer, M.P.; Rüdiger, S.G.D. Hsp90 Breaks the Deadlock of the Hsp70 Chaperone System. Mol. Cell 2018, 70, 545–552.e9. [Google Scholar]
- Johnson, B.D.; Schumacher, R.J.; Ross, E.D.; Toft, D.O. Hop modulates Hsp70/Hsp90 interactions in protein folding. J. Biol. Chem. 1998, 273, 3679–3686. [Google Scholar] [CrossRef] [Green Version]
- Schumacher, R.J.; Hansen, W.J.; Freeman, B.C.; Alnemri, E.; Litwack, G.; Toft, D.O. Cooperative action of Hsp70, Hsp90, and DnaJ proteins in protein renaturation. Biochemistry 1996, 35, 14889–14898. [Google Scholar] [CrossRef] [PubMed]
- Wegele, H.; Wandinger, S.K.; Schmid, A.B.; Reinstein, J.; Buchner, J. Substrate transfer from the chaperone Hsp70 to Hsp90. J. Mol. Biol. 2006, 356, 802–811. [Google Scholar] [CrossRef] [PubMed]
- Wayne, N.; Bolon, D.N. Dimerization of Hsp90 is required for in vivo function. Design and analysis of monomers and dimers. J. Biol. Chem. 2007, 282, 35386–35395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schopf, F.H.; Biebl, M.M.; Buchner, J. The HSP90 chaperone machinery. Nat. Rev. Mol. Cell Biol. 2017, 18, 345–360. [Google Scholar] [CrossRef]
- Taipale, M.; Tucker, G.; Peng, J.; Krykbaeva, I.; Lin, Z.-Y.; Larsen, B.; Choi, H.; Berger, B.; Gingras, A.-C.; Lindquist, S. A quantitative chaperone interaction network reveals the architecture of cellular protein homeostasis pathways. Cell 2014, 158, 434–448. [Google Scholar] [CrossRef] [Green Version]
- Taipale, M.; Krykbaeva, I.; Koeva, M.; Kayatekin, C.; Westover, K.D.; Karras, G.I.; Lindquist, S. Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell 2012, 150, 987–1001. [Google Scholar] [CrossRef] [Green Version]
- Jakob, U.; Lilie, H.; Meyer, I.; Buchner, J. Transient interaction of Hsp90 with early unfolding intermediates of citrate synthase. Implications for heat shock in vivo. J. Biol. Chem. 1995, 270, 7288–7294. [Google Scholar] [CrossRef] [Green Version]
- Karagöz, G.E.; Duarte, A.M.S.; Akoury, E.; Ippel, H.; Biernat, J.; Morán Luengo, T.; Radli, M.; Didenko, T.; Nordhues, B.A.; Veprintsev, D.B.; et al. Hsp90-Tau complex reveals molecular basis for specificity in chaperone action. Cell 2014, 156, 963–974. [Google Scholar] [CrossRef] [Green Version]
- Flynn, J.M.; Rossouw, A.; Cote-Hammarlof, P.; Fragata, I.; Mavor, D.; Hollins, C.; Bank, C.; Bolon, D.N. Comprehensive fitness maps of Hsp90 show widespread environmental dependence. Elife 2020, 9, e53810. [Google Scholar] [CrossRef]
- Gasch, A.P.; Spellman, P.T.; Kao, C.M.; Carmel-Harel, O.; Eisen, M.B.; Storz, G.; Botstein, D.; Brown, P.O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 2000, 11, 4241–4257. [Google Scholar] [CrossRef]
- Anckar, J.; Sistonen, L. Regulation of HSF1 function in the heat stress response: Implications in aging and disease. Annu. Rev. Biochem. 2011, 80, 1089–1115. [Google Scholar] [CrossRef] [PubMed]
- Triandafillou, C.G.; Katanski, C.D.; Dinner, A.R.; Drummond, D.A. Transient intracellular acidification regulates the core transcriptional heat shock response. bioRxiv 2019, 414706. [Google Scholar] [CrossRef]
- Abravaya, K.; Myers, M.P.; Murphy, S.P.; Morimoto, R.I. The human heat shock protein hsp70 interacts with HSF, the transcription factor that regulates heat shock gene expression. Genes Dev. 1992, 6, 1153–1164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baler, R.; Welch, W.J.; Voellmy, R. Heat shock gene regulation by nascent polypeptides and denatured proteins: Hsp70 as a potential autoregulatory factor. J. Cell Biol. 1992, 117, 1151–1159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, J.; Guo, Y.; Guettouche, T.; Smith, D.F.; Voellmy, R. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell 1998, 94, 471–480. [Google Scholar] [CrossRef] [Green Version]
- Albert, B.; Kos-Braun, I.C.; Henras, A.K.; Dez, C.; Rueda, M.P.; Zhang, X.; Gadal, O.; Kos, M.; Shore, D. A ribosome assembly stress response regulates transcription to maintain proteome homeostasis. eLife 2019, 8, e45002. [Google Scholar] [CrossRef]
- Masser, A.E.; Kang, W.; Roy, J.; Mohanakrishnan Kaimal, J.; Quintana-Cordero, J.; Friedländer, M.R.; Andréasson, C. Cytoplasmic protein misfolding titrates Hsp70 to activate nuclear Hsf1. eLife 2019, 8, e47791. [Google Scholar] [CrossRef]
- Kmiecik, S.W.; le Breton, L.; Mayer, M.P. Feedback regulation of heat shock factor 1 (Hsf1) activity by Hsp70-mediated trimer unzipping and dissociation from DNA. EMBO J. 2020, n/a, e104096. [Google Scholar] [CrossRef]
- Hahn, J.-S.; Hu, Z.; Thiele, D.J.; Iyer, V.R. Genome-Wide Analysis of the biology of stress responses through heat shock transcription factor. Mol. Cell. Biol. 2004, 24, 5249–5256. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; Chen, C.; Stevenson, M.A.; Auron, P.E.; Calderwood, S.K. Heat shock factor 1 represses transcription of theIL-1β gene through physical interaction with the nuclear factor of interleukin 6. J. Biol. Chem. 2002, 277, 11802–11810. [Google Scholar] [CrossRef] [Green Version]
- Espinet, C.; de la Torre, M.A.; Aldea, M.; Herrero, E. An efficient method to isolate yeast genes causing overexpression-mediated growth arrest. Yeast 1995, 11, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Dickey, C.A.; Kamal, A.; Lundgren, K.; Klosak, N.; Bailey, R.M.; Dunmore, J.; Ash, P.; Shoraka, S.; Zlatkovic, J.; Eckman, C.B.; et al. The high-affinity HSP90-CHIP complex recognizes and selectively degrades phosphorylated tau client proteins. J. Clin. Invest. 2007, 117, 648–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keller, J.A.; Simon, L.D. Divergent effects of a dnaK mutation on abnormal protein degradation in Escherichia coli. Mol. Microbiol. 1988, 2, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Sherman, M.Y.; Goldberg, A.L. Involvement of the chaperonin dnaK in the rapid degradation of a mutant protein in Escherichia coli. EMBO J. 1992, 11, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Straus, D.B.; Walter, W.A.; Gross, C.A. Escherichia coli heat shock gene mutants are defective in proteolysis. Genes Dev. 1988, 2, 1851–1858. [Google Scholar] [CrossRef]
- Wagner, I.; Arlt, H.; van Dyck, L.; Langer, T.; Neupert, W. Molecular chaperones cooperate with PIM1 protease in the degradation of misfolded proteins in mitochondria. EMBO J. 1994, 13, 5135–5145. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.H.; Sherman, M.Y.; Goldberg, A.L. The requirements of yeast Hsp70 of SSA family for the ubiquitin-dependent degradation of short-lived and abnormal proteins. Biochem. Biophys. Res. Commun. 2016, 475, 100–106. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Janich, S.; Cohn, J.A.; Wilson, J.M. The common variant of cystic fibrosis transmembrane conductance regulator is recognized by hsp70 and degraded in a pre-Golgi nonlysosomal compartment. Proc. Natl. Acad. Sci. USA 1993, 90, 9480–9484. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Nijbroek, G.; Sullivan, M.L.; McCracken, A.A.; Watkins, S.C.; Michaelis, S.; Brodsky, J.L. Hsp70 molecular chaperone facilitates endoplasmic reticulum-associated protein degradation of cystic fibrosis transmembrane conductance regulator in yeast. Mol. Biol. Cell 2001, 12, 1303–1314. [Google Scholar] [CrossRef] [Green Version]
- Bercovich, B.; Stancovski, I.; Mayer, A.; Blumenfeld, N.; Laszlo, A.; Schwartz, A.L.; Ciechanover, A. Ubiquitin-dependent degradation of certain protein substrates in vitro requires the molecular chaperone Hsc70. J. Biol. Chem. 1997, 272, 9002–9010. [Google Scholar] [CrossRef] [Green Version]
- Ehrlich, E.S.; Wang, T.; Luo, K.; Xiao, Z.; Niewiadomska, A.M.; Martinez, T.; Xu, W.; Neckers, L.; Yu, X.-F. Regulation of Hsp90 client proteins by a Cullin5-RING E3 ubiquitin ligase. Proc. Natl. Acad. Sci. USA 2009, 106, 20330–20335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerriero, C.J.; Weiberth, K.F.; Brodsky, J.L. Hsp70 targets a cytoplasmic quality control substrate to the San1p ubiquitin ligase. J. Biol. Chem. 2013, 288, 18506–18520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murata, S.; Minami, Y.; Minami, M.; Chiba, T.; Tanaka, K. CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Rep. 2001, 2, 1133–1138. [Google Scholar] [CrossRef]
- Park, S.-H.; Bolender, N.; Eisele, F.; Kostova, Z.; Takeuchi, J.; Coffino, P.; Wolf, D.H. The cytoplasmic Hsp70 chaperone machinery subjects misfolded and endoplasmic reticulum import-incompetent proteins to degradation via the ubiquitin-proteasome system. Mol. Biol. Cell 2007, 18, 153–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, R.; Chen, S.; Feldman, R.; Schieltz, D.; Yates, J.; Dohmen, J.; Deshaies, R.J. Proteasomal proteomics: Identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell 2000, 11, 3425–3439. [Google Scholar] [CrossRef] [Green Version]
- McClellan, A.J.; Scott, M.D.; Frydman, J. Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell 2005, 121, 739–748. [Google Scholar] [CrossRef] [Green Version]
- Schneider, C.; Sepp-Lorenzino, L.; Nimmesgern, E.; Ouerfelli, O.; Danishefsky, S.; Rosen, N.; Hartl, F.U. Pharmacologic shifting of a balance between protein refolding and degradation mediated by Hsp90. Proc. Natl. Acad. Sci. USA 1996, 93, 14536–14541. [Google Scholar] [CrossRef] [Green Version]
- Kelley, W.L. The J-domain family and the recruitment of chaperone power. Trends Biochem. Sci. 1998, 23, 222–227. [Google Scholar] [CrossRef]
- Kityk, R.; Kopp, J.; Mayer, M.P. Molecular mechanism of j-domain-triggered ATP hydrolysis by Hsp70 Chaperones. Mol. Cell 2018, 69, 227–237.e4. [Google Scholar] [CrossRef]
- Liu, Q.; Liang, C.; Zhou, L. Structural and functional analysis of the Hsp70/Hsp40 chaperone system. Protein Sci. 2020, 29, 378–390. [Google Scholar] [CrossRef]
- Qiu, X.-B.; Shao, Y.-M.; Miao, S.; Wang, L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. 2006, 63, 2560–2570. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Qian, X.; Sha, B. The crystal structure of the yeast Hsp40 Ydj1 complexed with its peptide substrate. Structure 2003, 11, 1475–1483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Rossi, P.; Kalodimos, C.G. Structural basis for client recognition and activity of Hsp40 chaperones. Science 2019, 365, 1313–1319. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Hendershot, L.M. ERdj3, a stress-inducible endoplasmic reticulum DnaJ homologue, serves as a CoFactor for BiP’s interactions with unfolded substrates. Mol. Biol. Cell 2005, 16, 40–50. [Google Scholar] [CrossRef] [Green Version]
- Perales-Calvo, J.; Muga, A.; Moro, F. Role of DnaJ G/F-rich domain in conformational recognition and binding of protein substrates. J. Biol. Chem. 2010, 285, 34231–34239. [Google Scholar] [CrossRef] [Green Version]
- Hageman, J.; Rujano, M.A.; van Waarde, M.A.W.H.; Kakkar, V.; Dirks, R.P.; Govorukhina, N.; Oosterveld-Hut, H.M.J.; Lubsen, N.H.; Kampinga, H.H. A DNAJB chaperone subfamily with HDAC-dependent activities suppresses toxic protein aggregation. Mol. Cell 2010, 37, 355–369. [Google Scholar] [CrossRef]
- den Brave, F.; Cairo, L.V.; Jagadeesan, C.; Ruger-Herreros, C.; Mogk, A.; Bukau, B.; Jentsch, S. Chaperone-mediated protein disaggregation triggers proteolytic clearance of intra-nuclear protein inclusions. Cell Rep. 2020, 31, 107680. [Google Scholar] [CrossRef]
- Samant, R.S.; Livingston, C.M.; Sontag, E.M.; Frydman, J. Distinct proteostasis circuits cooperate in nuclear and cytoplasmic protein quality control. Nature 2018, 563, 407–411. [Google Scholar] [CrossRef]
- Fang, N.N.; Chan, G.T.; Zhu, M.; Comyn, S.A.; Persaud, A.; Deshaies, R.J.; Rotin, D.; Gsponer, J.; Mayor, T. Rsp5/Nedd4 is the main ubiquitin ligase that targets cytosolic misfolded proteins following heat stress. Nat. Cell Biol. 2014, 16, 1227–1237. [Google Scholar] [CrossRef]
- Fang, N.N.; Zhu, M.; Rose, A.; Wu, K.-P.; Mayor, T. Deubiquitinase activity is required for the proteasomal degradation of misfolded cytosolic proteins upon heat-stress. Nat. Commun. 2016, 7, 12907. [Google Scholar] [CrossRef] [Green Version]
- Gaur, D.; Singh, P.; Guleria, J.; Gupta, A.; Kaur, S.; Sharma, D. The Yeast Hsp70 Co-chaperone Ydj1 regulates functional distinction of Ssa Hsp70s in the Hsp90 chaperoning pathway. Genetics 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez-González, C.; Lin, S.; Arkan, S.; Hansen, C. Co-chaperones DNAJA1 and DNAJB6 are critical for regulation of polyglutamine aggregation. Sci. Rep. 2020, 10, 8130. [Google Scholar] [CrossRef] [PubMed]
- Kim Chiaw, P.; Hantouche, C.; Wong, M.J.H.; Matthes, E.; Robert, R.; Hanrahan, J.W.; Shrier, A.; Young, J.C. Hsp70 and DNAJA2 limit CFTR levels through degradation. PLoS ONE 2019, 14, e0220984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, J.K.; Hutt, D.M.; Tait, B.; Guy, N.C.; Sivils, J.C.; Ortiz, N.R.; Payan, A.N.; Komaragiri, S.K.; Owens, J.J.; Culbertson, D.; et al. Management of Hsp90-Dependent protein folding by small molecules targeting the Aha1 Co-Chaperone. Cell Chem. Biol. 2020, 27, 292–305.e6. [Google Scholar] [CrossRef]
- Sahasrabudhe, P.; Rohrberg, J.; Biebl, M.M.; Rutz, D.A.; Buchner, J. The plasticity of the Hsp90 co-chaperone system. Mol. Cell 2017, 67, 947–961.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bengoechea, R.; Findlay, A.; Bhadra, A.; Shao, H.; Stein, K.; Pittman, S.; Daw, J.; Gestwicki, J.E.; True, H.; Weihl, C.C. Inhibition of DNAJ-HSP70 interaction improves strength in muscular dystrophy. bioRxiv 2020. [Google Scholar] [CrossRef]
- Bracher, A.; Verghese, J. The nucleotide exchange factors of Hsp70 molecular chaperones. Front. Mol. Biosci. 2015, 2. [Google Scholar] [CrossRef]
- Harrison, C. GrpE, a nucleotide exchange factor for DnaK. Cell Stress Chaperones 2003, 8, 218–224. [Google Scholar] [CrossRef] [Green Version]
- Harrison, C.J.; Hayer-Hartl, M.; Liberto, M.D.; Hartl, F.-U.; Kuriyan, J. Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science 1997, 276, 431–435. [Google Scholar] [CrossRef]
- Wu, C.-C.; Naveen, V.; Chien, C.-H.; Chang, Y.-W.; Hsiao, C.-D. Crystal structure of DnaK protein complexed with nucleotide exchange factor GrpE in DnaK chaperone system. J. Biol. Chem. 2012, 287, 21461–21470. [Google Scholar] [CrossRef] [Green Version]
- Gowda, N.K.C.; Kaimal, J.M.; Kityk, R.; Daniel, C.; Liebau, J.; Öhman, M.; Mayer, M.P.; Andréasson, C. Nucleotide exchange factors Fes1 and HspBP1 mimic substrate to release misfolded proteins from Hsp70. Nat. Struct. Mol. Biol. 2018, 25, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Shomura, Y.; Dragovic, Z.; Chang, H.-C.; Tzvetkov, N.; Young, J.C.; Brodsky, J.L.; Guerriero, V.; Hartl, F.U.; Bracher, A. Regulation of Hsp70 function by HspBP1: Structural analysis reveals an alternate mechanism for Hsp70 nucleotide exchange. Mol. Cell 2005, 17, 367–379. [Google Scholar] [PubMed]
- Gowda, N.K.C.; Kandasamy, G.; Froehlich, M.S.; Dohmen, R.J.; Andréasson, C. Hsp70 nucleotide exchange factor Fes1 is essential for ubiquitin-dependent degradation of misfolded cytosolic proteins. Proc. Natl. Acad. Sci. USA 2013, 110, 5975–5980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Easton, D.P.; Kaneko, Y.; Subjeck, J.R. The Hsp110 and Grp170 stress proteins: Newly recognized relatives of the Hsp70s. Cell Stress Chaperones 2000, 5, 276–290. [Google Scholar] [CrossRef] [Green Version]
- Gozzi, G.J.; Gonzalez, D.; Boudesco, C.; Dias, A.M.M.; Gotthard, G.; Uyanik, B.; Dondaine, L.; Marcion, G.; Hermetet, F.; Denis, C.; et al. Selecting the first chemical molecule inhibitor of HSP110 for colorectal cancer therapy. Cell Death Differ. 2020, 27, 117–129. [Google Scholar] [CrossRef]
- Liu, Q.; Hendrickson, W.A. Insights into Hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell 2007, 131, 106–120. [Google Scholar] [CrossRef] [Green Version]
- Andréasson, C.; Fiaux, J.; Rampelt, H.; Mayer, M.P.; Bukau, B. Hsp110 is a nucleotide-activated exchange factor for Hsp70. J. Biol. Chem. 2008, 283, 8877–8884. [Google Scholar] [CrossRef] [Green Version]
- Polier, S.; Dragovic, Z.; Hartl, F.U.; Bracher, A. Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell 2008, 133, 1068–1079. [Google Scholar] [CrossRef] [Green Version]
- Schuermann, J.P.; Jiang, J.; Cuellar, J.; Llorca, O.; Wang, L.; Gimenez, L.E.; Jin, S.; Taylor, A.B.; Demeler, B.; Morano, K.A.; et al. Structure of the Hsp110:Hsc70 nucleotide exchange machine. Mol. Cell 2008, 31, 232–243. [Google Scholar] [CrossRef] [Green Version]
- Raviol, H.; Sadlish, H.; Rodriguez, F.; Mayer, M.P.; Bukau, B. Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J. 2006, 25, 2510–2518. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Sarbeng, E.B.; Vorvis, C.; Kumar, D.P.; Zhou, L.; Liu, Q. Unique peptide substrate binding properties of 110-kDa heat-shock protein (Hsp110) determine its distinct chaperone activity. J. Biol. Chem. 2012, 287, 5661–5672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serlidaki, D.; van Waarde, M.A.W.H.; Rohland, L.; Wentink, A.S.; Dekker, S.L.; Kamphuis, M.J.; Boertien, J.M.; Brunsting, J.F.; Nillegoda, N.B.; Bukau, B.; et al. Functional diversity between HSP70 paralogs due to variable interactions with specific co-chaperones. J. Biol. Chem. 2020, 295, 7301–7316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takayama, S.; Xie, Z.; Reed, J.C. An Evolutionarily conserved family of Hsp70/Hsc70 Molecular chaperone regulators. J. Biol. Chem. 1999, 274, 781–786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sondermann, H.; Scheufler, C.; Schneider, C.; Höhfeld, J.; Hartl, F.-U.; Moarefi, I. Structure of a Bag/Hsc70 complex: Convergent functional evolution of Hsp70 nucleotide exchange factors. Science 2001, 291, 1553–1557. [Google Scholar] [CrossRef]
- Mariotto, E.; Viola, G.; Zanon, C.; Aveic, S. A BAG’s life: Every connection matters in cancer. Pharmacol. Ther. 2020, 209, 107498. [Google Scholar] [CrossRef]
- Tanaka, H.; Takahashi, T.; Xie, Y.; Minami, R.; Yanagi, Y.; Hayashishita, M.; Suzuki, R.; Yokota, N.; Shimada, M.; Mizushima, T.; et al. A conserved island of BAG6/Scythe is related to ubiquitin domains and participates in short hydrophobicity recognition. FEBS J. 2016, 283, 662–677. [Google Scholar] [CrossRef] [Green Version]
- Kriegenburg, F.; Jakopec, V.; Poulsen, E.G.; Nielsen, S.V.; Roguev, A.; Krogan, N.; Gordon, C.; Fleig, U.; Hartmann-Petersen, R. A Chaperone-assisted degradation pathway targets kinetochore proteins to ensure genome stability. PLoS Genet. 2014, 10, e1004140. [Google Scholar] [CrossRef]
- Poulsen, E.G.; Kampmeyer, C.; Kriegenburg, F.; Johansen, J.V.; Hofmann, K.; Holmberg, C.; Hartmann-Petersen, R. UBL/BAG-domain co-chaperones cause cellular stress upon overexpression through constitutive activation of Hsf1. Cell Stress Chaperones 2017, 22, 143–154. [Google Scholar] [CrossRef] [Green Version]
- Vjestica, A.; Zhang, D.; Liu, J.; Oliferenko, S. Hsp70-Hsp40 chaperone complex functions in controlling polarized growth by repressing Hsf1-driven heat stress-associated transcription. PLoS Genet. 2013, 9, e1003886. [Google Scholar] [CrossRef] [Green Version]
- Sroka, K.; Voigt, A.; Deeg, S.; Reed, J.C.; Schulz, J.B.; Bähr, M.; Kermer, P. BAG1 modulates huntingtin toxicity, aggregation, degradation, and subcellular distribution. J. Neurochem. 2009, 111, 801–807. [Google Scholar] [CrossRef]
- Tsukahara, F.; Maru, Y. Bag1 directly routes immature BCR-ABL for proteasomal degradation. Blood 2010, 116, 3582–3592. [Google Scholar] [CrossRef] [PubMed]
- Hantouche, C.; Williamson, B.; Valinsky, W.C.; Solomon, J.; Shrier, A.; Young, J.C. Bag1 Co-chaperone promotes TRC8 E3 ligase-dependent degradation of misfolded human ether a Go-Go-related gene (hERG) Potassium Channels. J. Biol. Chem. 2017, 292, 2287–2300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ballinger, C.A.; Connell, P.; Wu, Y.; Hu, Z.; Thompson, L.J.; Yin, L.-Y.; Patterson, C. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol. 1999, 19, 4535–4545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quintana-Gallardo, L.; Martín-Benito, J.; Marcilla, M.; Espadas, G.; Sabidó, E.; Valpuesta, J.M. The cochaperone CHIP marks Hsp70- and Hsp90-bound substrates for degradation through a very flexible mechanism. Sci. Rep. 2019, 9, 5102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lüders, J.; Demand, J.; Höhfeld, J. The Ubiquitin-related BAG-1 Provides a Link between the Molecular Chaperones Hsc70/Hsp70 and the Proteasome. J. Biol. Chem. 2000, 275, 4613–4617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saeki, Y.; Sone, T.; Toh-e, A.; Yokosawa, H. Identification of ubiquitin-like protein-binding subunits of the 26S proteasome. Biochem. Biophys. Res. Commun. 2002, 296, 813–819. [Google Scholar] [CrossRef]
- Alberti, S.; Demand, J.; Esser, C.; Emmerich, N.; Schild, H.; Höhfeld, J. Ubiquitylation of BAG-1 suggests a novel regulatory mechanism during the sorting of chaperone substrates to the proteasome. J. Biol. Chem. 2002, 277, 45920–45927. [Google Scholar] [CrossRef] [Green Version]
- Gässler, C.S.; Wiederkehr, T.; Brehmer, D.; Bukau, B.; Mayer, M.P. Bag-1M accelerates nucleotide release for human Hsc70 and Hsp70 and can act concentration-dependent as positive and negative cofactor. J. Biol. Chem. 2001, 276, 32538–32544. [Google Scholar] [CrossRef] [Green Version]
- Höhfeld, J.; Jentsch, S. GrpE-like regulation of the Hsc70 chaperone by the anti-apoptotic protein BAG-1. EMBO J. 1997, 16, 6209–6216. [Google Scholar] [CrossRef] [Green Version]
- Diamant, S.; Goloubinoff, P. Temperature-controlled activity of DnaK-DnaJ-GrpE chaperones: Protein-folding arrest and recovery during and after heat shock depends on the substrate protein and the GrpE concentration. Biochemistry 1998, 37, 9688–9694. [Google Scholar] [CrossRef]
- Irmer, H.; Höhfeld, J. Characterization of functional domains of the eukaryotic Co-chaperone hip. J. Biol. Chem. 1997, 272, 2230–2235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nollen, E.A.A.; Kabakov, A.E.; Brunsting, J.F.; Kanon, B.; Höhfeld, J.; Kampinga, H.H. Modulation of in vivo HSP70 chaperone activity by hip and Bag-1. J. Biol. Chem. 2001, 276, 4677–4682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Hartl, F.U.; Bracher, A. Structure and function of Hip, an attenuator of the Hsp70 chaperone cycle. Nat. Struct. Mol. Biol. 2013, 20, 929–935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alberti, S.; Böhse, K.; Arndt, V.; Schmitz, A.; Höhfeld, J. The cochaperone HspBP1 inhibits the CHIP ubiquitin ligase and stimulates the maturation of the cystic fibrosis transmembrane conductance regulator. Mol. Biol. Cell 2004, 15, 4003–4010. [Google Scholar] [CrossRef] [PubMed]
- Esser, C.; Alberti, S.; Höhfeld, J. Cooperation of molecular chaperones with the ubiquitin/proteasome system. Biochim. Biophys. Acta 2004, 1695, 171–188. [Google Scholar] [CrossRef] [Green Version]
- Imai, Y.; Soda, M.; Hatakeyama, S.; Akagi, T.; Hashikawa, T.; Nakayama, K.-I.; Takahashi, R. CHIP Is associated with parkin, a gene responsible for familial parkinson’s disease, and enhances its ubiquitin ligase activity. Mol. Cell 2002, 10, 55–67. [Google Scholar] [CrossRef]
- Aguileta, M.A.; Korac, J.; Durcan, T.M.; Trempe, J.-F.; Haber, M.; Gehring, K.; Elsasser, S.; Waidmann, O.; Fon, E.A.; Husnjak, K. The E3 ubiquitin ligase parkin is recruited to the 26 S proteasome via the proteasomal ubiquitin receptor rpn13. J. Biol. Chem. 2015, 290, 7492–7505. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; Marcu, M.; Yuan, X.; Mimnaugh, E.; Patterson, C.; Neckers, L. Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc. Natl. Acad. Sci. USA 2002, 99, 12847–12852. [Google Scholar] [CrossRef] [Green Version]
- Theodoraki, M.A.; Nillegoda, N.B.; Saini, J.; Caplan, A.J. A network of ubiquitin ligases is important for the dynamics of misfolded protein aggregates in yeast. J. Biol. Chem. 2012, 287, 23911–23922. [Google Scholar] [CrossRef] [Green Version]
- Mariappan, M.; Li, X.; Stefanovic, S.; Sharma, A.; Mateja, A.; Keenan, R.J.; Hegde, R.S. A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature 2010, 466, 1120–1124. [Google Scholar] [CrossRef] [Green Version]
- Rodrigo-Brenni, M.C.; Gutierrez, E.; Hegde, R.S. Cytosolic quality control of mislocalized proteins requires RNF126 recruitment to Bag6. Mol. Cell 2014, 55, 227–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Liu, Y.; Soetandyo, N.; Baek, K.; Hegde, R.; Ye, Y. A ubiquitin ligase-associated chaperone holdase maintains polypeptides in soluble states for proteasome degradation. Mol. Cell 2011, 42, 758–770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minami, R.; Hayakawa, A.; Kagawa, H.; Yanagi, Y.; Yokosawa, H.; Kawahara, H. BAG-6 is essential for selective elimination of defective proteasomal substrates. J. Cell Biol. 2010, 190, 637–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, K.; Hayashishita, M.; Minami, S.; Suzuki, K.; Hagiwara, T.; Noguchi, A.; Kawahara, H. Elimination of a signal sequence-uncleaved form of defective HLA protein through BAG6. Sci. Rep. 2017, 7, 1–13. [Google Scholar]
- Hessa, T.; Sharma, A.; Mariappan, M.; Eshleman, H.D.; Gutierrez, E.; Hegde, R.S. Protein targeting and degradation are coupled for elimination of mislocalized proteins. Nature 2011, 475, 394–397. [Google Scholar] [CrossRef]
- Kadowaki, H.; Nagai, A.; Maruyama, T.; Takami, Y.; Satrimafitrah, P.; Kato, H.; Honda, A.; Hatta, T.; Natsume, T.; Sato, T.; et al. Pre-emptive quality control protects the ER from protein overload via the proximity of ERAD components and SRP. Cell Rep. 2015, 13, 944–956. [Google Scholar] [CrossRef] [Green Version]
- Ganji, R.; Mukkavalli, S.; Somanji, F.; Raman, M. The VCP-UBXN1 complex mediates triage of ubiquitylated cytosolic proteins bound to the BAG6 complex. Mol. Cell. Biol. 2018, 38, e00154-18. [Google Scholar] [CrossRef] [Green Version]
- Kuwabara, N.; Minami, R.; Yokota, N.; Matsumoto, H.; Senda, T.; Kawahara, H.; Kato, R. Structure of a BAG6 (Bcl-2-associated Athanogene 6)-Ubl4a (Ubiquitin-like Protein 4a) complex reveals a novel binding interface that functions in tail-anchored protein biogenesis. J. Biol. Chem. 2015, 290, 9387–9398. [Google Scholar] [CrossRef] [Green Version]
- Mock, J.-Y.; Chartron, J.W.; Zaslaver, M.; Xu, Y.; Ye, Y.; Clemons, W.M. Bag6 complex contains a minimal tail-anchor-targeting module and a mock BAG domain. Proc. Natl. Acad. Sci. USA 2015, 112, 106–111. [Google Scholar] [CrossRef] [Green Version]
- Kawahara, H.; Minami, R.; Yokota, N. BAG6/BAT3: Emerging roles in quality control for nascent polypeptides. J. Biochem. 2013, 153, 147–160. [Google Scholar] [CrossRef] [Green Version]
- Mock, J.-Y.; Xu, Y.; Ye, Y.; Clemons, W.M. Structural basis for regulation of the nucleo-cytoplasmic distribution of Bag6 by TRC35. Proc. Natl. Acad. Sci. USA 2017, 114, 11679–11684. [Google Scholar] [CrossRef] [Green Version]
- Shao, S.; Rodrigo-Brenni, M.C.; Kivlen, M.H.; Hegde, R.S. Mechanistic basis for a molecular triage reaction. Science 2017, 355, 298–302. [Google Scholar] [CrossRef] [Green Version]
- Stefanovic, S.; Hegde, R.S. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell 2007, 128, 1147–1159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Cai, M.; Yang, Y.; Huang, L.; Ye, Y. SGTA recognizes a noncanonical ubiquitin-like domain in the Bag6-Ubl4A-Trc35 complex to promote endoplasmic reticulum-associated degradation. Cell Rep. 2012, 2, 1633–1644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, F.; Brown, E.C.; Mak, G.; Zhuang, J.; Denic, V. A chaperone cascade sorts proteins for posttranslational membrane insertion into the endoplasmic reticulum. Mol. Cell 2010, 40, 159–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, X.; Wang, L.; Wang, Y.; Ji, J.; Li, J.; Wang, Z.; Li, C.; Zhang, Y.; Zhang, Z.-R. RNF126-mediated reubiquitination is required for proteasomal degradation of p97-extracted membrane proteins. Mol. Cell 2020, 79, 320–331.e9. [Google Scholar] [CrossRef]
- Kikukawa, Y.; Minami, R.; Shimada, M.; Kobayashi, M.; Tanaka, K.; Yokosawa, H.; Kawahara, H. Unique proteasome subunit Xrpn10c is a specific receptor for the antiapoptotic ubiquitin-like protein Scythe. FEBS J. 2005, 272, 6373–6386. [Google Scholar] [CrossRef]
- Collins, G.A.; Goldberg, A.L. Proteins containing ubiquitin-like (Ubl) domains not only bind to 26S proteasomes but also induce their activation. Proc. Natl. Acad. Sci. USA 2020, 117, 4664–4674. [Google Scholar] [CrossRef]
- Liou, S.-T.; Cheng, M.-Y.; Wang, C. SGT2 and MDY2 interact with molecular chaperone YDJ1 in Saccharomyces cerevisiae. Cell Stress Chaperones 2007, 12, 59–70. [Google Scholar] [CrossRef]
- Leznicki, P.; Korac-Prlic, J.; Kliza, K.; Husnjak, K.; Nyathi, Y.; Dikic, I.; High, S. Binding of SGTA to Rpn13 selectively modulates protein quality control. J. Cell Sci. 2015, 128, 3187–3196. [Google Scholar] [CrossRef] [Green Version]
- Liou, S.-T.; Wang, C. Small glutamine-rich tetratricopeptide repeat-containing protein is composed of three structural units with distinct functions. Arch. Biochem. Biophys. 2005, 435, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Thapaliya, A.; Nyathi, Y.; Martínez-Lumbreras, S.; Krysztofinska, E.M.; Evans, N.J.; Terry, I.L.; High, S.; Isaacson, R.L. SGTA interacts with the proteasomal ubiquitin receptor Rpn13 via a carboxylate clamp mechanism. Sci. Rep. 2016, 6, 36622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kandasamy, G.; Andréasson, C. Hsp70-Hsp110 chaperones deliver ubiquitin-dependent and -independent substrates to the 26S proteasome for proteolysis in yeast. J. Cell Sci. 2018, 131, jcs210948. [Google Scholar] [CrossRef] [Green Version]
- Mandal, A.K.; Gibney, P.A.; Nillegoda, N.B.; Theodoraki, M.A.; Caplan, A.J.; Morano, K.A. Hsp110 chaperones control client fate determination in the Hsp70–Hsp90 chaperone system. Mol. Biol. Cell 2010, 21, 1439–1448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenbaum, J.C.; Fredrickson, E.K.; Oeser, M.L.; Garrett-Engele, C.M.; Locke, M.N.; Richardson, L.A.; Nelson, Z.W.; Hetrick, E.D.; Milac, T.I.; Gottschling, D.E.; et al. Disorder targets misorder in nuclear quality control degradation: A disordered ubiquitin ligase directly recognizes its misfolded substrates. Mol. Cell 2011, 41, 93–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boomsma, W.; Nielsen, S.V.; Lindorff-Larsen, K.; Hartmann-Petersen, R.; Ellgaard, L. Bioinformatics analysis identifies several intrinsically disordered human E3 ubiquitin-protein ligases. PeerJ 2016, 4, e1725. [Google Scholar] [CrossRef] [Green Version]
- Jones, R.D.; Enam, C.; Ibarra, R.; Borror, H.R.; Mostoller, K.E.; Fredrickson, E.K.; Lin, J.; Chuang, E.; March, Z.; Shorter, J.; et al. The extent of Ssa1/Ssa2 Hsp70 chaperone involvement in nuclear protein quality control degradation varies with the substrate. Mol. Biol. Cell 2020, 31, 221–233. [Google Scholar] [CrossRef]
- Geffen, Y.; Appleboim, A.; Gardner, R.G.; Friedman, N.; Sadeh, R.; Ravid, T. Mapping the landscape of a eukaryotic degronome. Mol. Cell 2016, 63, 1055–1065. [Google Scholar] [CrossRef] [Green Version]
- Kim, I.; Miller, C.R.; Young, D.L.; Fields, S. High-throughput analysis of in vivo protein stability. Mol. Cell. Proteom. 2013, 12, 3370–3378. [Google Scholar] [CrossRef] [Green Version]
- Arteaga, M.F.; Wang, L.; Ravid, T.; Hochstrasser, M.; Canessa, C.M. An amphipathic helix targets serum and glucocorticoid-induced kinase 1 to the endoplasmic reticulum-associated ubiquitin-conjugation machinery. Proc. Natl. Acad. Sci. USA 2006, 103, 11178–11183. [Google Scholar] [CrossRef] [Green Version]
- Fredrickson, E.K.; Rosenbaum, J.C.; Locke, M.N.; Milac, T.I.; Gardner, R.G. Exposed hydrophobicity is a key determinant of nuclear quality control degradation. Mol. Biol. Cell 2011, 22, 2384–2395. [Google Scholar] [CrossRef]
- Banerjee, R.; Jayaraj, G.G.; Peter, J.J.; Kumar, V.; Mapa, K. Monitoring conformational heterogeneity of the lid of DnaK substrate-binding domain during its chaperone cycle. FEBS J. 2016, 283, 2853–2868. [Google Scholar] [CrossRef]
- Medicherla, B.; Kostova, Z.; Schaefer, A.; Wolf, D.H. A genomic screen identifies Dsk2p and Rad23p as essential components of ER-associated degradation. EMBO Rep. 2004, 5, 692–697. [Google Scholar] [CrossRef] [Green Version]
- Kaye, F.J.; Modi, S.; Ivanovska, I.; Koonin, E.V.; Thress, K.; Kubo, A.; Kornbluth, S.; Rose, M.D. A family of ubiquitin-like proteins binds the ATPase domain of Hsp70-like Stch. FEBS Lett. 2000, 467, 348–355. [Google Scholar] [CrossRef] [Green Version]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abildgaard, A.B.; Gersing, S.K.; Larsen-Ledet, S.; Nielsen, S.V.; Stein, A.; Lindorff-Larsen, K.; Hartmann-Petersen, R. Co-Chaperones in Targeting and Delivery of Misfolded Proteins to the 26S Proteasome. Biomolecules 2020, 10, 1141. https://0-doi-org.brum.beds.ac.uk/10.3390/biom10081141
Abildgaard AB, Gersing SK, Larsen-Ledet S, Nielsen SV, Stein A, Lindorff-Larsen K, Hartmann-Petersen R. Co-Chaperones in Targeting and Delivery of Misfolded Proteins to the 26S Proteasome. Biomolecules. 2020; 10(8):1141. https://0-doi-org.brum.beds.ac.uk/10.3390/biom10081141
Chicago/Turabian StyleAbildgaard, Amanda B., Sarah K. Gersing, Sven Larsen-Ledet, Sofie V. Nielsen, Amelie Stein, Kresten Lindorff-Larsen, and Rasmus Hartmann-Petersen. 2020. "Co-Chaperones in Targeting and Delivery of Misfolded Proteins to the 26S Proteasome" Biomolecules 10, no. 8: 1141. https://0-doi-org.brum.beds.ac.uk/10.3390/biom10081141