Highly Specialized Ubiquitin-Like Modifications: Shedding Light into the UFM1 Enigma
Abstract
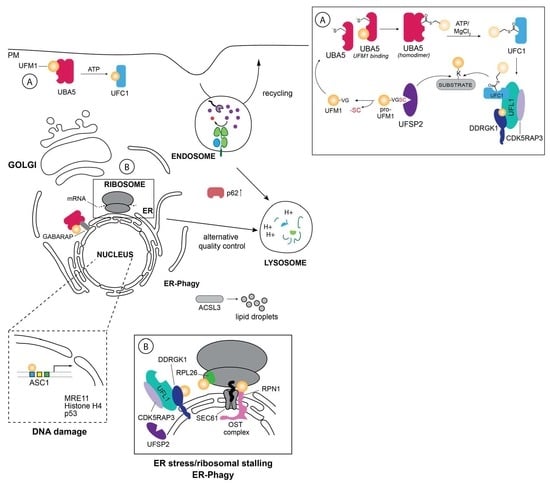
:1. Introduction
2. The UFM1 (de)Conjugation System
2.1. UBA5

2.2. UFC1
2.3. UFL1
2.4. UFSP1 and UFSP2
3. Biological Function of UFMylation

3.1. UFM1—A Matter of Survival during Embryonic Development
3.2. ER Stress: UFM1 to the Rescue
3.3. Autophagy—Self Eating for Survival
3.4. Transcriptional Regulation
3.5. Signaling Pathways
3.6. UFMylation and the DNA Damage Response
4. UFMylation and Disease
5. Tools to Study UFMylation
5.1. Identifying UFM1 Substrates—The Key to Unraveling UFM1 Biology
| Method | Constructs and Experimental Conditions | Substrates or Interacting Proteins | Reference |
|---|---|---|---|
| Affinity purification | Strep-Tag UFM1 overexpressed in insulin-producing MIN6 cells | UBA5, UFC1, UFL1, DDRGK1 | [33] |
| GST-UFM1 | |||
| Flag-His-UFM1 | ASC1 | [34] | |
| Flag-UFL1 and DDRGK1 | p53 | [58] | |
| His6-UFM1ΔSC | RPL26 and RPL26L1 | [35,36] | |
| Flag-UFM1 (WT and ΔC3) in UFSP2 CRISPR knockout cells and mass spec | |||
| Flag-DDRGK1 | ANT3 1, NUP160 1, and SF3B1 1 (DDRGK1 associated proteins) | [76] | |
| BirAOPT-UFM1-UFC1 vector | 20 UFM1 substrates/interactors found incl. MRE11. Only CYB5R3 and PSMB5 were validated | [125] | |
| StrepII-tagged UFM1 | RPL26, RPS3 1, RPS20 1, RPL10 1 and several UFM1 interactors | [81] | |
| Yeast two-hybrid screening | Originally GATE16 was discovered as a UBA5 interactor, and later identification of UFM1 by AP-MS | [15] | |
| Yeast two hybrid and PTM reconstitution in E. coli | pDEST32 bait and prey vectors; pYESS-PIP and pYESSPTM vectors | 16 potential substrates or interactors; only DDRGK1, MT1M1, and TSC22D3 1 were validated | [126] |
| Co-immunoprecipitation and microscopy experiments | His-UFM1 pulldown under denaturing conditions after induction of DNA damage followed by MS/MS | Histone H4 and three other potential substrates (ZNF281 1, ZIC2 1, and CAPNS1 1) | [39] |
| Colocalization of UFL1 and y-H2AX upon irradiation; co-immunoprecipitation of HA-UFL1 revealed interaction with MRE11 | MRE11 | [40] | |
| Protein microarray and ELISA | GST-NCAM140 (cytoplasmic domain) | UFC1 interaction with the cytoplasmic domain of NCAM 140 | [91] |
| Protein microarray | Profiling of Ubl modification of about 9000 proteins before and after release from mitotic arrest | Identification of 202 UFM1 targets (transmembrane transporters, ion channels, cytokine transporters); hit validation required | [55] |
| Peptide array | UBA5 peptides and GST-LC3 and GST-GABARAP | Mapping of LIR/UFIM motif in the C-terminal domain of UBA5 | [19] |
| Peptide-competition coupled affinity proteomics screen | IP-MS screen under ER stress conditions using a synthetic ATG8-interacting motif peptide with higher affinity than ATG8 | C53 (or CDK5RAP3) was revealed to bind ATG8 | [68] |
| CRISPR screens | CRISPR knockout screens (CRISPRi) | p62 (SQSTM), DDRGK1, RPN1 | [38,59] |
| CRISPR-mediated HA-tagging of endogenous proteins (hATG8) | ACSL3 | [56] | |
| Chromatin Co-immunoprecipitation (ChIP) | Endogenous and overexpressed XBP-1s (Flag-XBP-1s) | XBP-1s binds to the promoter region of UFM1 | [54] |
| Bioinformatic analysis | Analysis of proteins with UFM1 binding motif [19] | STK38 (NDR1) binding to UFMylated Histone H4 in complex with KAP-1 1 and SUV39H1 | [104] |
5.2. Towards Development of a UFM1 Toolkit Enabling Inhibitor Discovery
6. Outlook and Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Kim, W.; Bennett, E.J.; Huttlin, E.L.; Guo, A.; Li, J.; Possemato, A.; Sowa, M.E.; Rad, R.; Rush, J.; Comb, M.J.; et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 2011, 44, 325–340. [Google Scholar] [CrossRef] [Green Version]
- Hendriks, I.A.; Vertegaal, A.C. Label-Free Identification and Quantification of SUMO Target Proteins. Methods Mol. Biol. 2016, 1475, 171–193. [Google Scholar] [CrossRef] [PubMed]
- .Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muratani, M.; Tansey, W.P. How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 2003, 4, 192–201. [Google Scholar] [CrossRef]
- Kernan, J.; Bonacci, T.; Emanuele, M.J. Who guards the guardian? Mechanisms that restrain APC/C during the cell cycle. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 1924–1933. [Google Scholar] [CrossRef] [PubMed]
- Hicke, L. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2001, 2, 195–201. [Google Scholar] [CrossRef]
- McCann, A.P.; Scott, C.J.; van Schaeybroeck, S.; Burrows, J.F. Deubiquitylating enzymes in receptor endocytosis and trafficking. Biochem. J. 2016, 473, 4507–4525. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.P.; Durocher, D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell 2013, 49, 795–807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, Y.T.; Ciechanover, A. The Ubiquitin Code in the Ubiquitin-Proteasome System and Autophagy. Trends Biochem. Sci. 2017, 42, 873–886. [Google Scholar] [CrossRef]
- Gerakis, Y.; Quintero, M.; Li, H.; Hetz, C. The UFMylation System in Proteostasis and Beyond. Trends Cell Biol. 2019, 29, 974–986. [Google Scholar] [CrossRef]
- Fang, Z.; Pan, Z. Essential Role of Ubiquitin-Fold Modifier 1 Conjugation in DNA Damage Response. DNA Cell Biol. 2019, 38, 1030–1039. [Google Scholar] [CrossRef]
- Varshavsky, A. The Ubiquitin System, Autophagy, and Regulated Protein Degradation. Annu. Rev. Biochem. 2017, 86, 123–128. [Google Scholar] [CrossRef]
- Daniel, J.; Liebau, E. The ufm1 cascade. Cells 2014, 3, 627–638. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Kumar, M.; Wiener, R. Decrypting UFMylation: How Proteins Are Modified with UFM1. Biomolecules 2020, 10, 1442. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, M.; Chiba, T.; Tatsumi, K.; Iemura, S.; Tanida, I.; Okazaki, N.; Ueno, T.; Kominami, E.; Natsume, T.; Tanaka, K. A novel protein-conjugating system for Ufm1, a ubiquitin-fold modifier. EMBO J. 2004, 23, 1977–1986. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.; Xu, X. UFMylation: A Unique & Fashionable Modification for Life. Genom. Proteom. Bioinform. 2016, 14, 140–146. [Google Scholar] [CrossRef] [Green Version]
- Schulman, B.A.; Harper, J.W. Ubiquitin-like protein activation by E1 enzymes: The apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 2009, 10, 319–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Padala, P.; Oweis, W.; Mashahreh, B.; Soudah, N.; Cohen-Kfir, E.; Todd, E.A.; Berndsen, C.E.; Wiener, R. Novel insights into the interaction of UBA5 with UFM1 via a UFM1-interacting sequence. Sci. Rep. 2017, 7, 508. [Google Scholar] [CrossRef] [Green Version]
- Habisov, S.; Huber, J.; Ichimura, Y.; Akutsu, M.; Rogova, N.; Loehr, F.; McEwan, D.G.; Johansen, T.; Dikic, I.; Doetsch, V.; et al. Structural and Functional Analysis of a Novel Interaction Motif within UFM1-activating Enzyme 5 (UBA5) Required for Binding to Ubiquitin-like Proteins and Ufmylation. J. Biol. Chem. 2016, 291, 9025–9041. [Google Scholar] [CrossRef] [Green Version]
- Soudah, N.; Padala, P.; Hassouna, F.; Kumar, M.; Mashahreh, B.; Lebedev, A.A.; Isupov, M.N.; Cohen-Kfir, E.; Wiener, R. An N-Terminal Extension to UBA5 Adenylation Domain Boosts UFM1 Activation: Isoform-Specific Differences in Ubiquitin-like Protein Activation. J. Mol. Biol. 2019, 431, 463–478. [Google Scholar] [CrossRef] [Green Version]
- Lv, Z.; Olsen, S.K. UFM1-Activating Enzyme 5 (Uba5) Requires an Extension to Get the Job Done Right. J. Mol. Biol. 2019, 431, 479–482. [Google Scholar] [CrossRef] [PubMed]
- Bacik, J.P.; Walker, J.R.; Ali, M.; Schimmer, A.D.; Dhe-Paganon, S. Crystal structure of the human ubiquitin-activating enzyme 5 (UBA5) bound to ATP: Mechanistic insights into a minimalistic E1 enzyme. J. Biol. Chem. 2010, 285, 20273–20280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gavin, J.M.; Hoar, K.; Xu, Q.; Ma, J.; Lin, Y.; Chen, J.; Chen, W.; Bruzzese, F.J.; Harrison, S.; Mallender, W.D.; et al. Mechanistic study of Uba5 enzyme and the Ufm1 conjugation pathway. J. Biol. Chem. 2014, 289, 22648–22658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mashahreh, B.; Hassouna, F.; Soudah, N.; Cohen-Kfir, E.; Strulovich, R.; Haitin, Y.; Wiener, R. Trans-binding of UFM1 to UBA5 stimulates UBA5 homodimerization and ATP binding. FASEB J. 2018, 32, 2794–2802. [Google Scholar] [CrossRef] [Green Version]
- Huber, J.; Obata, M.; Gruber, J.; Akutsu, M.; Lohr, F.; Rogova, N.; Guntert, P.; Dikic, I.; Kirkin, V.; Komatsu, M.; et al. An atypical LIR motif within UBA5 (ubiquitin like modifier activating enzyme 5) interacts with GABARAP proteins and mediates membrane localization of UBA5. Autophagy 2020, 16, 256–270. [Google Scholar] [CrossRef]
- Ishimura, R.; Obata, M.; Kageyama, S.; Daniel, J.; Tanaka, K.; Komatsu, M. A novel approach to assess the ubiquitin-fold modifier 1-system in cells. FEBS Lett. 2017, 591, 196–204. [Google Scholar] [CrossRef] [Green Version]
- Mizushima, T.; Tatsumi, K.; Ozaki, Y.; Kawakami, T.; Suzuki, A.; Ogasahara, K.; Komatsu, M.; Kominami, E.; Tanaka, K.; Yamane, T. Crystal structure of Ufc1, the Ufm1-conjugating enzyme. Biochem. Biophys. Res. Commun. 2007, 362, 1079–1084. [Google Scholar] [CrossRef]
- Liu, G.; Aramini, J.; Atreya, H.S.; Eletsky, A.; Xiao, R.; Acton, T.; Ma, L.; Montelione, G.T.; Szyperski, T. GFT NMR based resonance assignment for the 21 kDa human protein UFC1. J. Biomol. NMR 2005, 32, 261. [Google Scholar] [CrossRef]
- Liu, G.; Forouhar, F.; Eletsky, A.; Atreya, H.S.; Aramini, J.M.; Xiao, R.; Huang, Y.J.; Abashidze, M.; Seetharaman, J.; Liu, J.; et al. NMR and X-RAY structures of human E2-like ubiquitin-fold modifier conjugating enzyme 1 (UFC1) reveal structural and functional conservation in the metazoan UFM1-UBA5-UFC1 ubiquination pathway. J. Struct. Funct. Genom. 2009, 10, 127–136. [Google Scholar] [CrossRef]
- Xie, S. Characterization, crystallization and preliminary X-ray crystallographic analysis of the human Uba5 C-terminus-Ufc1 complex. Acta Crystallogr. F Struct. Biol. Commun. 2014, 70, 1093–1097. [Google Scholar] [CrossRef] [Green Version]
- Xie, Z.; Fang, Z.; Pan, Z. Ufl1/RCAD, a Ufm1 E3 ligase, has an intricate connection with ER stress. Int. J. Biol. Macromol. 2019, 135, 760–767. [Google Scholar] [CrossRef]
- Tatsumi, K.; Sou, Y.S.; Tada, N.; Nakamura, E.; Iemura, S.; Natsume, T.; Kang, S.H.; Chung, C.H.; Kasahara, M.; Kominami, E.; et al. A novel type of E3 ligase for the Ufm1 conjugation system. J. Biol. Chem. 2010, 285, 5417–5427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemaire, K.; Moura, R.F.; Granvik, M.; Igoillo-Esteve, M.; Hohmeier, H.E.; Hendrickx, N.; Newgard, C.B.; Waelkens, E.; Cnop, M.; Schuit, F. Ubiquitin fold modifier 1 (UFM1) and its target UFBP1 protect pancreatic beta cells from ER stress-induced apoptosis. PLoS ONE 2011, 6, e18517. [Google Scholar] [CrossRef] [Green Version]
- Yoo, H.M.; Kang, S.H.; Kim, J.Y.; Lee, J.E.; Seong, M.W.; Lee, S.W.; Ka, S.H.; Sou, Y.S.; Komatsu, M.; Tanaka, K.; et al. Modification of ASC1 by UFM1 is crucial for ERalpha transactivation and breast cancer development. Mol. Cell 2014, 56, 261–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walczak, C.P.; Leto, D.E.; Zhang, L.; Riepe, C.; Muller, R.Y.; DaRosa, P.A.; Ingolia, N.T.; Elias, J.E.; Kopito, R.R. Ribosomal protein RPL26 is the principal target of UFMylation. Proc. Natl. Acad. Sci. USA 2019, 116, 1299–1308. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Xu, Y.; Rogers, H.; Saidi, L.; Noguchi, C.T.; Li, H.; Yewdell, J.W.; Guydosh, N.R.; Ye, Y. UFMylation of RPL26 links translocation-associated quality control to endoplasmic reticulum protein homeostasis. Cell Res. 2020, 30, 5–20. [Google Scholar] [CrossRef]
- Scheel, H.; Hofmann, K. Prediction of a common structural scaffold for proteasome lid, COP9-signalosome and eIF3 complexes. BMC Bioinform. 2005, 6, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, J.R.; Lingeman, E.; Luong, T.; Ahmed, S.; Muhar, M.; Nguyen, T.; Olzmann, J.A.; Corn, J.E. A Genome-wide ER-phagy Screen Highlights Key Roles of Mitochondrial Metabolism and ER-Resident UFMylation. Cell 2020, 180, 1160–1177. [Google Scholar] [CrossRef]
- Qin, B.; Yu, J.; Nowsheen, S.; Wang, M.; Tu, X.; Liu, T.; Li, H.; Wang, L.; Lou, Z. UFL1 promotes histone H4 ufmylation and ATM activation. Nat. Commun. 2019, 10, 1242. [Google Scholar] [CrossRef]
- Wang, Z.; Gong, Y.; Peng, B.; Shi, R.; Fan, D.; Zhao, H.; Zhu, M.; Zhang, H.; Lou, Z.; Zhou, J.; et al. MRE11 UFMylation promotes ATM activation. Nucleic Acids Res. 2019, 47, 4124–4135. [Google Scholar] [CrossRef] [Green Version]
- Duda, D.M.; Borg, L.A.; Scott, D.C.; Hunt, H.W.; Hammel, M.; Schulman, B.A. Structural insights into NEDD8 activation of cullin-RING ligases: Conformational control of conjugation. Cell 2008, 134, 995–1006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mak, G.W.; Lai, W.L.; Zhou, Y.; Li, M.; Ng, I.O.; Ching, Y.P. CDK5RAP3 is a novel repressor of p14ARF in hepatocellular carcinoma cells. PLoS ONE 2012, 7, e42210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, H.; Wu, J.; He, C.; Yang, W.; Li, H. Tumor suppressor protein C53 antagonizes checkpoint kinases to promote cyclin-dependent kinase 1 activation. Cell Res. 2009, 19, 458–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, H.; Luo, S.; Li, H. Cdk5 activator-binding protein C53 regulates apoptosis induced by genotoxic stress via modulating the G2/M DNA damage checkpoint. J. Biol. Chem. 2005, 280, 20651–20659. [Google Scholar] [CrossRef] [Green Version]
- Kwon, J.; Cho, H.J.; Han, S.H.; No, J.G.; Kwon, J.Y.; Kim, H. A novel LZAP-binding protein, NLBP, inhibits cell invasion. J. Biol. Chem. 2010, 285, 12232–12240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiwaku, H.; Yoshimura, N.; Tamura, T.; Sone, M.; Ogishima, S.; Watase, K.; Tagawa, K.; Okazawa, H. Suppression of the novel ER protein Maxer by mutant ataxin-1 in Bergman glia contributes to non-cell-autonomous toxicity. EMBO J. 2010, 29, 2446–2460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Lei, G.; Mei, M.; Tang, Y.; Li, H. A novel C53/LZAP-interacting protein regulates stability of C53/LZAP and DDRGK domain-containing Protein 1 (DDRGK1) and modulates NF-kappaB signaling. J. Biol. Chem. 2010, 285, 15126–15136. [Google Scholar] [CrossRef] [Green Version]
- Yang, R.; Wang, H.; Kang, B.; Chen, B.; Shi, Y.; Yang, S.; Sun, L.; Liu, Y.; Xiao, W.; Zhang, T.; et al. CDK5RAP3, a UFL1 substrate adaptor, is crucial for liver development. Development 2019, 146, dev169235. [Google Scholar] [CrossRef] [Green Version]
- Kang, S.H.; Kim, G.R.; Seong, M.; Baek, S.H.; Seol, J.H.; Bang, O.S.; Ovaa, H.; Tatsumi, K.; Komatsu, M.; Tanaka, K.; et al. Two novel ubiquitin-fold modifier 1 (Ufm1)-specific proteases, UfSP1 and UfSP2. J. Biol. Chem. 2007, 282, 5256–5262. [Google Scholar] [CrossRef] [Green Version]
- Ha, B.H.; Ahn, H.C.; Kang, S.H.; Tanaka, K.; Chung, C.H.; Kim, E.E. Structural basis for Ufm1 processing by UfSP1. J. Biol. Chem. 2008, 283, 14893–14900. [Google Scholar] [CrossRef] [Green Version]
- Ha, B.H.; Jeon, Y.J.; Shin, S.C.; Tatsumi, K.; Komatsu, M.; Tanaka, K.; Watson, C.M.; Wallis, G.; Chung, C.H.; Kim, E.E. Structure of ubiquitin-fold modifier 1-specific protease UfSP2. J. Biol. Chem. 2011, 286, 10248–10257. [Google Scholar] [CrossRef] [Green Version]
- Witting, K.F.; van Noort, G.J.v.d.H.; Kofoed, C.; Ormeno, C.T.; Atmioui, D.E.; Mulder, M.P.C.; Ovaa, H. Generation of the UFM1 Toolkit for Profiling UFM1-Specific Proteases and Ligases. Angew. Chem. Int. Ed. Engl. 2018, 57, 14164–14168. [Google Scholar] [CrossRef]
- Cai, Y.; Singh, N.; Li, H. Essential role of Ufm1 conjugation in the hematopoietic system. Exp. Hematol. 2016, 44, 442–446. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Zhang, M.; Wu, J.; Lei, G.; Li, H. Transcriptional regulation of the Ufm1 conjugation system in response to disturbance of the endoplasmic reticulum homeostasis and inhibition of vesicle trafficking. PLoS ONE 2012, 7, e48587. [Google Scholar] [CrossRef] [Green Version]
- Merbl, Y.; Refour, P.; Patel, H.; Springer, M.; Kirschner, M.W. Profiling of ubiquitin-like modifications reveals features of mitotic control. Cell 2013, 152, 1160–1172. [Google Scholar] [CrossRef] [Green Version]
- Eck, F.; Phuyal, S.; Smith, M.D.; Kaulich, M.; Wilkinson, S.; Farhan, H.; Behrends, C. ACSL3 is a novel GABARAPL2 interactor that links ufmylation and lipid droplet biogenesis. J. Cell Sci. 2020, 133, jcs243477. [Google Scholar] [CrossRef]
- Lin, J.X.; Xie, X.S.; Weng, X.F.; Qiu, S.L.; Yoon, C.; Lian, N.Z.; Xie, J.W.; Wang, J.B.; Lu, J.; Chen, Q.Y.; et al. UFM1 suppresses invasive activities of gastric cancer cells by attenuating the expres7sion of PDK1 through PI3K/AKT signaling. J. Exp. Clin. Cancer Res. 2019, 38, 410. [Google Scholar] [CrossRef]
- Liu, J.; Guan, D.; Dong, M.; Yang, J.; Wei, H.; Liang, Q.; Song, L.; Xu, L.; Bai, J.; Liu, C.; et al. UFMylation maintains tumour suppressor p53 stability by antagonizing its ubiquitination. Nat. Cell Biol. 2020, 22, 1056–1063. [Google Scholar] [CrossRef]
- De Jesus, R.; Moretti, F.; McAllister, G.; Wang, Z.; Bergman, P.; Liu, S.; Frias, E.; Alford, J.; Reece-Hoyes, J.S.; Lindeman, A.; et al. Functional CRISPR screening identifies the ufmylation pathway as a regulator of SQSTM1/p62. Elife 2016, 5, e17290. [Google Scholar] [CrossRef]
- Tatsumi, K.; Yamamoto-Mukai, H.; Shimizu, R.; Waguri, S.; Sou, Y.S.; Sakamoto, A.; Taya, C.; Shitara, H.; Hara, T.; Chung, C.H.; et al. The Ufm1-activating enzyme Uba5 is indispensable for erythroid differentiation in mice. Nat. Commun. 2011, 2, 181. [Google Scholar] [CrossRef]
- Cai, Y.; Pi, W.; Sivaprakasam, S.; Zhu, X.; Zhang, M.; Chen, J.; Makala, L.; Lu, C.; Wu, J.; Teng, Y.; et al. UFBP1, a Key Component of the Ufm1 Conjugation System, Is Essential for Ufmylation-Mediated Regulation of Erythroid Development. PLoS Genet. 2015, 11, e1005643. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhu, X.; Zhang, Y.; Cai, Y.; Chen, J.; Sivaprakasam, S.; Gurav, A.; Pi, W.; Makala, L.; Wu, J.; et al. RCAD/Ufl1, a Ufm1 E3 ligase, is essential for hematopoietic stem cell function and murine hematopoiesis. Cell Death Differ. 2015, 22, 1922–1934. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, Y.; Song, L.; Zeng, L.; Yi, W.; Liu, T.; Chen, H.; Wang, M.; Ju, Z.; Cong, Y.S. A critical role of DDRGK1 in endoplasmic reticulum homoeostasis via regulation of IRE1alpha stability. Nat. Commun. 2017, 8, 14186. [Google Scholar] [CrossRef]
- Cai, Y.; Zhu, G.; Liu, S.; Pan, Z.; Quintero, M.; Poole, C.J.; Lu, C.; Zhu, H.; Islam, B.; Riggelen, J.V.; et al. Indispensable role of the Ubiquitin-fold modifier 1-specific E3 ligase in maintaining intestinal homeostasis and controlling gut inflammation. Cell Discov. 2019, 5, 7. [Google Scholar] [CrossRef]
- Lin, K.Y.; Kao, S.H.; Lai, C.M.; Chen, C.T.; Wu, C.Y.; Hsu, H.J.; Wang, W.D. Tumor Suppressor Lzap Suppresses Wnt/beta-Catenin Signaling to Promote Zebrafish Embryonic Ventral Cell Fates via the Suppression of Inhibitory Phosphorylation of Glycogen Synthase Kinase 3. J. Biol. Chem. 2015, 290, 29808–29819. [Google Scholar] [CrossRef] [Green Version]
- Bruce, A.E.E.; Heisenberg, C.P. Mechanisms of zebrafish epiboly: A current view. Curr. Top. Dev. Biol. 2020, 136, 319–341. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Sicinski, P. A kinase of many talents: Non-neuronal functions of CDK5 in development and disease. Open Biol. 2020, 10, 190287. [Google Scholar] [CrossRef] [Green Version]
- Stephani, M.; Picchianti, L.; Dagdas, Y. C53 is a cross-kingdom conserved reticulophagy receptor that bridges the gap betweenselective autophagy and ribosome stalling at the endoplasmic reticulum. Autophagy 2020, 1–2. [Google Scholar] [CrossRef]
- Ching, Y.P.; Qi, Z.; Wang, J.H. Cloning of three novel neuronal Cdk5 activator binding proteins. Gene 2000, 242, 285–294. [Google Scholar] [CrossRef]
- Neziri, D.; Ilhan, A.; Maj, M.; Majdic, O.; Baumgartner-Parzer, S.; Cohen, G.; Base, W.; Wagner, L. Cloning and molecular characterization of Dashurin encoded by C20orf116, a PCI-domain containing protein. Biochim. Biophys. Acta 2010, 1800, 430–438. [Google Scholar] [CrossRef]
- Hetz, C. The unfolded protein response: Controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 2012, 13, 89–102. [Google Scholar] [CrossRef]
- Chen, X.; Karnovsky, A.; Sans, M.D.; Andrews, P.C.; Williams, J.A. Molecular characterization of the endoplasmic reticulum: Insights from proteomic studies. Proteomics 2010, 10, 4040–4052. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, Y.; Yin, Y.; Gao, G.; Li, S.; Jiang, Y.; Gu, X.; Luo, J. SPD—A web-based secreted protein database. Nucleic Acids Res. 2005, 33, D169–D173. [Google Scholar] [CrossRef] [Green Version]
- Rashid, H.O.; Yadav, R.K.; Kim, H.R.; Chae, H.J. ER stress: Autophagy induction, inhibition and selection. Autophagy 2015, 11, 1956–1977. [Google Scholar] [CrossRef]
- Azfer, A.; Niu, J.; Rogers, L.M.; Adamski, F.M.; Kolattukudy, P.E. Activation of endoplasmic reticulum stress response during the development of ischemic heart disease. Am. J. Physiol. Heart Circ. Physiol. 2006, 291, H1411–H1420. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Lei, Q.; Li, D.; Zhang, Y.; Jiang, X.; Hu, Z.; Xu, G. Proteomic and Biochemical Analyses Reveal a Novel Mechanism for Promoting Protein Ubiquitination and Degradation by UFBP1, a Key Component of Ufmylation. J. Proteome Res. 2018, 17, 1509–1520. [Google Scholar] [CrossRef]
- Sun, S.; Shi, G.; Sha, H.; Ji, Y.; Han, X.; Shu, X.; Ma, H.; Inoue, T.; Gao, B.; Kim, H.; et al. IRE1alpha is an endogenous substrate of endoplasmic-reticulum-associated degradation. Nat. Cell Biol. 2015, 17, 1546–1555. [Google Scholar] [CrossRef] [Green Version]
- Bagola, K.; Mehnert, M.; Jarosch, E.; Sommer, T. Protein dislocation from the ER. Biochim. Biophys. Acta 2011, 1808, 925–936. [Google Scholar] [CrossRef] [Green Version]
- Simsek, D.; Tiu, G.C.; Flynn, R.A.; Byeon, G.W.; Leppek, K.; Xu, A.F.; Chang, H.Y.; Barna, M. The Mammalian Ribo-interactome Reveals Ribosome Functional Diversity and Heterogeneity. Cell 2017, 169, 1051–1065. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Jia, M.; Su, M.; Hu, X.; Li, J.; Xu, Y.; Qiu, W. Ufmylation Is Activated in Renal Cancer and Is Not Associated with von Hippel-Lindau Mutation. DNA Cell Biol. 2020, 39, 654–660. [Google Scholar] [CrossRef]
- Schuren, A.B.C.; Boer, I.G.J.; Bouma, E.M.; Van de Weijer, M.L.; Costa, A.I.; Hubel, P.; Pichlmair, A.; Lebbink, R.J.; Wiertz, E. The UFM1 Pathway Impacts HCMV US2-Mediated Degradation of HLA Class I. Molecules 2021, 26, 287. [Google Scholar] [CrossRef] [PubMed]
- Dikic, I.; Elazar, Z. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 349–364. [Google Scholar] [CrossRef]
- Hu, X.; Zhang, H.; Song, Y.; Zhuang, L.; Yang, Q.; Pan, M.; Chen, F. Ubiquitin fold modifier 1 activates NF-kappaB pathway by down-regulating LZAP expression in the macrophage of diabetic mouse model. Biosci. Rep. 2020, 40, BSR20191672. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yue, G.; Ma, W.; Zhang, A.; Zou, J.; Cai, Y.; Tang, X.; Wang, J.; Liu, J.; Li, H.; et al. Ufm1-Specific Ligase Ufl1 Regulates Endoplasmic Reticulum Homeostasis and Protects Against Heart Failure. Circ. Heart Fail. 2018, 11, e004917. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.; Cai, Y.; Patton, T.; Graves, S.H.; Li, H.; Sabbatini, M.E. RCAD/BiP pathway is necessary for the proper synthesis of digestive enzymes and secretory function of the exocrine pancreas. Am. J. Physiol. Gastrointest. Liver Physiol. 2017, 312, G314–G326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johansen, T.; Lamark, T. Selective Autophagy: ATG8 Family Proteins, LIR Motifs and Cargo Receptors. J. Mol. Biol. 2020, 432, 80–103. [Google Scholar] [CrossRef]
- Stolz, A.; Ernst, A.; Dikic, I. Cargo recognition and trafficking in selective autophagy. Nat. Cell Biol. 2014, 16, 495–501. [Google Scholar] [CrossRef]
- Song, S.; Tan, J.; Miao, Y.; Zhang, Q. Crosstalk of ER stress-mediated autophagy and ER-phagy: Involvement of UPR and the core autophagy machinery. J. Cell Physiol. 2018, 233, 3867–3874. [Google Scholar] [CrossRef]
- Strzyz, P. Foundations of ER-phagy regulation. Nat. Rev. Mol. Cell Biol. 2020, 21, 251. [Google Scholar] [CrossRef]
- Jarc, E.; Petan, T. Lipid Droplets and the Management of Cellular Stress. Yale J. Biol. Med. 2019, 92, 435–452. [Google Scholar]
- Homrich, M.; Wobst, H.; Laurini, C.; Sabrowski, J.; Schmitz, B.; Diestel, S. Cytoplasmic domain of NCAM140 interacts with ubiquitin-fold modifier-conjugating enzyme-1 (Ufc1). Exp. Cell Res. 2014, 324, 192–199. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2020, 22, 96–118. [Google Scholar] [CrossRef]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef]
- Yu, T.; Shan, T.D.; Li, J.Y.; Huang, C.Z.; Wang, S.Y.; Ouyang, H.; Lu, X.J.; Xu, J.H.; Zhong, W.; Chen, Q.K. Knockdown of linc-UFC1 suppresses proliferation and induces apoptosis of colorectal cancer. Cell Death Dis. 2016, 7, e2228. [Google Scholar] [CrossRef]
- Beckedorff, F.C.; Amaral, M.S.; Deocesano-Pereira, C.; Verjovski-Almeida, S. Long non-coding RNAs and their implications in cancer epigenetics. Biosci. Rep. 2013, 33, 54. [Google Scholar] [CrossRef]
- Li, Y.Y.; Zhang, G.Y.; He, J.P.; Zhang, D.D.; Kong, X.X.; Yuan, H.M.; Chen, F.L. Ufm1 inhibits LPS-induced endothelial cell inflammatory responses through the NF-kappaB signaling pathway. Int. J. Mol. Med. 2017, 39, 1119–1126. [Google Scholar] [CrossRef] [Green Version]
- Kuang, M.; Yang, M.; Li, L.; Li, C.; Wang, G. UFM1-Specific Ligase 1 Ligating Enzyme 1 Mediates Milk Protein and Fat Synthesis-Related Gene Expression via the JNK Signaling Pathway in Mouse Mammary Epithelial Cells. Oxid. Med. Cell Longev. 2020, 2020, 4045674. [Google Scholar] [CrossRef]
- Lin, J.X.; Yoon, C.; Li, P.; Ryeom, S.W.; Cho, S.J.; Zheng, C.H.; Xie, J.W.; Wang, J.B.; Lu, J.; Chen, Q.Y.; et al. CDK5RAP3 as tumour suppressor negatively regulates self-renewal and invasion and is regulated by ERK1/2 signalling in human gastric cancer. Br. J. Cancer 2020, 123, 1131–1144. [Google Scholar] [CrossRef]
- Chen, Q.Y.; Liu, L.C.; Wang, J.B.; Xie, J.W.; Lin, J.X.; Lu, J.; Cao, L.L.; Lin, M.; Tu, R.H.; Huang, C.M.; et al. CDK5RAP3 Inhibits the Translocation of MCM6 to Influence the Prognosis in Gastric Cancer. J. Cancer 2019, 10, 4488–4498. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.X.; Xie, X.S.; Weng, X.F.; Zheng, C.H.; Xie, J.W.; Wang, J.B.; Lu, J.; Chen, Q.Y.; Cao, L.L.; Lin, M.; et al. Low expression of CDK5RAP3 and DDRGK1 indicates a poor prognosis in patients with gastric cancer. World J. Gastroenterol. 2018, 24, 3898–3907. [Google Scholar] [CrossRef]
- Yang, G.; Wang, Y.; Chen, Y.; Huang, R. UFL1 attenuates IL-1beta-induced inflammatory response in human osteoarthritis chondrocytes. Int. Immunopharmacol. 2020, 81, 106278. [Google Scholar] [CrossRef]
- Harper, J.W.; Elledge, S.J. The DNA damage response: Ten years after. Mol. Cell 2007, 28, 739–745. [Google Scholar] [CrossRef]
- Lamarche, B.J.; Orazio, N.I.; Weitzman, M.D. The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett. 2010, 584, 3682–3695. [Google Scholar] [CrossRef] [Green Version]
- Qin, B.; Yu, J.; Nowsheen, S.; Zhao, F.; Wang, L.; Lou, Z. STK38 promotes ATM activation by acting as a reader of histone H4 ufmylation. Sci. Adv. 2020, 6, eaax8214. [Google Scholar] [CrossRef]
- Dai, C.; Gu, W. p53 post-translational modification: Deregulated in tumorigenesis. Trends Mol. Med. 2010, 16, 528–536. [Google Scholar] [CrossRef] [Green Version]
- Nahorski, M.S.; Maddirevula, S.; Ishimura, R.; Alsahli, S.; Brady, A.F.; Begemann, A.; Mizushima, T.; Guzman-Vega, F.J.; Obata, M.; Ichimura, Y.; et al. Biallelic UFM1 and UFC1 mutations expand the essential role of ufmylation in brain development. Brain 2018, 141, 1934–1945. [Google Scholar] [CrossRef] [Green Version]
- Yu, L.; Li, G.; Deng, J.; Jiang, X.; Xue, J.; Zhu, Y.; Huang, W.; Tang, B.; Duan, R. The UFM1 cascade times mitosis entry associated with microcephaly. FASEB J. 2020, 34, 1319–1330. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, E.M.C.; Bertini, E.; Kalaydjieva, L.; Morar, B.; Dojcakova, D.; Liu, J.; Vanderver, A.; Curiel, J.; Persoon, C.M.; Diodato, D.; et al. UFM1 founder mutation in the Roma population causes recessive variant of H-ABC. Neurology 2017, 89, 1821–1828. [Google Scholar] [CrossRef] [Green Version]
- Colin, E.; Daniel, J.; Ziegler, A.; Wakim, J.; Scrivo, A.; Haack, T.B.; Khiati, S.; Denomme, A.S.; Amati-Bonneau, P.; Charif, M.; et al. Biallelic Variants in UBA5 Reveal that Disruption of the UFM1 Cascade Can Result in Early-Onset Encephalopathy. Am. J. Hum. Genet. 2016, 99, 695–703. [Google Scholar] [CrossRef] [Green Version]
- Muona, M.; Ishimura, R.; Laari, A.; Ichimura, Y.; Linnankivi, T.; Keski-Filppula, R.; Herva, R.; Rantala, H.; Paetau, A.; Poyhonen, M.; et al. Biallelic Variants in UBA5 Link Dysfunctional UFM1 Ubiquitin-like Modifier Pathway to Severe Infantile-Onset Encephalopathy. Am. J. Hum. Genet. 2016, 99, 683–694. [Google Scholar] [CrossRef] [Green Version]
- Duan, R.; Shi, Y.; Yu, L.; Zhang, G.; Li, J.; Lin, Y.; Guo, J.; Wang, J.; Shen, L.; Jiang, H.; et al. UBA5 Mutations Cause a New Form of Autosomal Recessive Cerebellar Ataxia. PLoS ONE 2016, 11, e0149039. [Google Scholar] [CrossRef]
- Cabrera-Serrano, M.; Coote, D.J.; Azmanov, D.; Goullee, H.; Andersen, E.; McLean, C.; Davis, M.; Ishimura, R.; Stark, Z.; Vallat, J.M.; et al. A homozygous UBA5 pathogenic variant causes a fatal congenital neuropathy. J. Med. Genet. 2020, 57, 835–842. [Google Scholar] [CrossRef]
- Liu, H.; Gong, M.; French, B.A.; Li, J.; Tillman, B.; French, S.W. Mallory-Denk Body (MDB) formation modulates Ufmylation expression epigenetically in alcoholic hepatitis (AH) and non-alcoholic steatohepatitis (NASH). Exp. Mol. Pathol. 2014, 97, 477–483. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Zhang, H.; Zhuang, L.; Jin, G.; Yang, Q.; Li, M.; Sun, W.; Chen, F. Ubiquitin-Fold Modifier-1 Participates in the Diabetic Inflammatory Response by Regulating NF-kappaB p65 Nuclear Translocation and the Ubiquitination and Degradation of IkappaBalpha. Drug Des. Dev. Ther. 2020, 14, 795–810. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.; Sun, Q.Q.; Liu, T.X.; Lu, K.; Zhang, N.; Zhu, Y.; Chen, M. Serum lncRNA-UFC1 as a potential biomarker for diagnosis and prognosis of pancreatic cancer. Int. J. Clin. Exp. Pathol. 2019, 12, 4125–4129. [Google Scholar]
- Xie, R.; Wang, M.; Zhou, W.; Wang, D.; Yuan, Y.; Shi, H.; Wu, L. Long Non-Coding RNA (LncRNA) UFC1/miR-34a Contributes to Proliferation and Migration in Breast Cancer. Med. Sci. Monit. 2019, 25, 7149–7157. [Google Scholar] [CrossRef]
- Zhang, X.; Liang, W.; Liu, J.; Zang, X.; Gu, J.; Pan, L.; Shi, H.; Fu, M.; Huang, Z.; Zhang, Y.; et al. Long non-coding RNA UFC1 promotes gastric cancer progression by regulating miR-498/Lin28b. J. Exp. Clin. Cancer Res. 2018, 37, 134. [Google Scholar] [CrossRef] [Green Version]
- Arnadottir, G.A.; Jensson, B.O.; Marelsson, S.E.; Sulem, G.; Oddsson, A.; Kristjansson, R.P.; Benonisdottir, S.; Gudjonsson, S.A.; Masson, G.; Thorisson, G.A.; et al. Compound heterozygous mutations in UBA5 causing early-onset epileptic encephalopathy in two sisters. BMC Med. Genet. 2017, 18, 103. [Google Scholar] [CrossRef] [Green Version]
- Daida, A.; Hamano, S.I.; Ikemoto, S.; Matsuura, R.; Nakashima, M.; Matsumoto, N.; Kato, M. Biallelic loss-of-function UBA5 mutations in a patient with intractable West syndrome and profound failure to thrive. Epileptic Disord. 2018, 20, 313–318. [Google Scholar] [CrossRef]
- Mignon-Ravix, C.; Milh, M.; Kaiser, C.S.; Daniel, J.; Riccardi, F.; Cacciagli, P.; Nagara, M.; Busa, T.; Liebau, E.; Villard, L. Abnormal function of the UBA5 protein in a case of early developmental and epileptic encephalopathy with suppression-burst. Hum. Mutat. 2018, 39, 934–938. [Google Scholar] [CrossRef] [PubMed]
- Low, K.J.; Baptista, J.; Babiker, M.; Caswell, R.; King, C.; Ellard, S.; Scurr, I. Hemizygous UBA5 missense mutation unmasks recessive disorder in a patient with infantile-onset encephalopathy, acquired microcephaly, small cerebellum, movement disorder and severe neurodevelopmental delay. Eur. J. Med. Genet. 2019, 62, 97–102. [Google Scholar] [CrossRef]
- Di Rocco, M.; Rusmini, M.; Caroli, F.; Madeo, A.; Bertamino, M.; Marre-Brunenghi, G.; Ceccherini, I. Novel spondyloepimetaphyseal dysplasia due to UFSP2 gene mutation. Clin. Genet. 2018, 93, 671–674. [Google Scholar] [CrossRef] [Green Version]
- Watson, C.M.; Crinnion, L.A.; Gleghorn, L.; Newman, W.G.; Ramesar, R.; Beighton, P.; Wallis, G.A. Identification of a mutation in the ubiquitin-fold modifier 1-specific peptidase 2 gene, UFSP2, in an extended South African family with Beukes hip dysplasia. S. Afr. Med. J. 2015, 105, 558–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, G.; Tang, S.; Wang, H.; Pan, H.; Zhang, W.; Huang, Y.; Kong, J.; Wang, Y.; Gu, J.; Wang, Y. UFSP2-related spondyloepimetaphyseal dysplasia: A confirmatory report. Eur. J. Med. Genet. 2020, 63, 104021. [Google Scholar] [CrossRef] [PubMed]
- Pirone, L.; Xolalpa, W.; Sigurethsson, J.O.; Ramirez, J.; Perez, C.; Gonzalez, M.; de Sabando, A.R.; Elortza, F.; Rodriguez, M.S.; Mayor, U.; et al. A comprehensive platform for the analysis of ubiquitin-like protein modifications using in vivo biotinylation. Sci. Rep. 2017, 7, 40756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Han, T.; Guo, R.; Chen, P.; Peng, C.; Prag, G.; Hu, R. An Integrative Synthetic Biology Approach to Interrogating Cellular Ubiquitin and Ufm Signaling. Int. J. Mol. Sci. 2020, 21, 4231. [Google Scholar] [CrossRef] [PubMed]
- Harder, Z.; Zunino, R.; McBride, H. Sumo1 conjugates mitochondrial substrates and participates in mitochondrial fission. Curr. Biol. 2004, 14, 340–345. [Google Scholar] [CrossRef]
- Voeltz, G.K.; Prinz, W.A.; Shibata, Y.; Rist, J.M.; Rapoport, T.A. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 2006, 124, 573–586. [Google Scholar] [CrossRef] [Green Version]
- Birchler, J.A.; Veitia, R.A. Gene balance hypothesis: Connecting issues of dosage sensitivity across biological disciplines. Proc. Natl. Acad. Sci. USA 2012, 109, 14746–14753. [Google Scholar] [CrossRef] [Green Version]
- Gibson, T.J.; Seiler, M.; Veitia, R.A. The transience of transient overexpression. Nat. Methods 2013, 10, 715–721. [Google Scholar] [CrossRef]
- Fulzele, A.; Bennett, E.J. Ubiquitin diGLY Proteomics as an Approach to Identify and Quantify the Ubiquitin-Modified Proteome. Methods Mol. Biol. 2018, 1844, 363–384. [Google Scholar] [CrossRef]
- Ogunkoya, A.O.; Pattabiraman, V.R.; Bode, J.W. Sequential alpha-ketoacid-hydroxylamine (KAHA) ligations: Synthesis of C-terminal variants of the modifier protein UFM1. Angew. Chem. Int. Ed. Engl. 2012, 51, 9693–9697. [Google Scholar] [CrossRef]
- Da Silva, S.R.; Paiva, S.L.; Bancerz, M.; Geletu, M.; Lewis, A.M.; Chen, J.; Cai, Y.; Lukkarila, J.L.; Li, H.; Gunning, P.T. A selective inhibitor of the UFM1-activating enzyme, UBA5. Bioorg. Med. Chem. Lett. 2016, 26, 4542–4547. [Google Scholar] [CrossRef]
- Roberts, A.M.; Miyamoto, D.K.; Huffman, T.R.; Bateman, L.A.; Ives, A.N.; Akopian, D.; Heslin, M.J.; Contreras, C.M.; Rape, M.; Skibola, C.F.; et al. Chemoproteomic Screening of Covalent Ligands Reveals UBA5 As a Novel Pancreatic Cancer Target. ACS Chem. Biol. 2017, 12, 899–904. [Google Scholar] [CrossRef]
- Chen, F.; Xing, C.; Zhang, W.; Li, J.; Hu, T.; Li, L.; Li, H.; Cai, Y. Salubrinal, a novel inhibitor of eIF-2alpha dephosphorylation, promotes erythropoiesis at early stage targeted by ufmylation pathway. J. Cell Physiol. 2019, 234, 18560–18570. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Gonzalez-Prieto, R.; Xiao, Z.; Verlaan-de Vries, M.; Vertegaal, A.C.O. The STUbL RNF4 regulates protein group SUMOylation by targeting the SUMO conjugation machinery. Nat. Commun. 2017, 8, 1809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salas-Lloret, D.; Agabitini, G.; Gonzalez-Prieto, R. TULIP2: An Improved Method for the Identification of Ubiquitin E3-Specific Targets. Front. Chem. 2019, 7, 802. [Google Scholar] [CrossRef] [PubMed]
- Low, T.Y.; Peng, M.; Magliozzi, R.; Mohammed, S.; Guardavaccaro, D.; Heck, A.J. A systems-wide screen identifies substrates of the SCFbetaTrCP ubiquitin ligase. Sci. Signal. 2014, 7, rs8. [Google Scholar] [CrossRef]
- Chen, D.; Liu, X.; Xia, T.; Tekcham, D.S.; Wang, W.; Chen, H.; Li, T.; Lu, C.; Ning, Z.; Liu, X.; et al. A Multidimensional Characterization of E3 Ubiquitin Ligase and Substrate Interaction Network. iScience 2019, 16, 177–191. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Xie, P.; Lu, L.; Wang, J.; Diao, L.; Liu, Z.; Guo, F.; He, Y.; Liu, Y.; Huang, Q.; et al. An integrated bioinformatics platform for investigating the human E3 ubiquitin ligase-substrate interaction network. Nat. Commun. 2017, 8, 347. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, V.N.; Huang, K.Y.; Weng, J.T.; Lai, K.R.; Lee, T.Y. UbiNet: An online resource for exploring the functional associations and regulatory networks of protein ubiquitylation. Database (Oxford) 2016, 2016, baw054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoo, H.M.; Park, J.H.; Jeon, Y.J.; Chung, C.H. Ubiquitin-fold modifier 1 acts as a positive regulator of breast cancer. Front. Endocrinol. (Lausanne) 2015, 6, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Witting, K.F.; Mulder, M.P.C. Highly Specialized Ubiquitin-Like Modifications: Shedding Light into the UFM1 Enigma. Biomolecules 2021, 11, 255. https://0-doi-org.brum.beds.ac.uk/10.3390/biom11020255
Witting KF, Mulder MPC. Highly Specialized Ubiquitin-Like Modifications: Shedding Light into the UFM1 Enigma. Biomolecules. 2021; 11(2):255. https://0-doi-org.brum.beds.ac.uk/10.3390/biom11020255
Chicago/Turabian StyleWitting, Katharina F., and Monique P.C. Mulder. 2021. "Highly Specialized Ubiquitin-Like Modifications: Shedding Light into the UFM1 Enigma" Biomolecules 11, no. 2: 255. https://0-doi-org.brum.beds.ac.uk/10.3390/biom11020255