The Molecular Basis of Human Anophthalmia and Microphthalmia
Abstract
:1. Introduction
2. The Genetic Basis of Human Eye Development
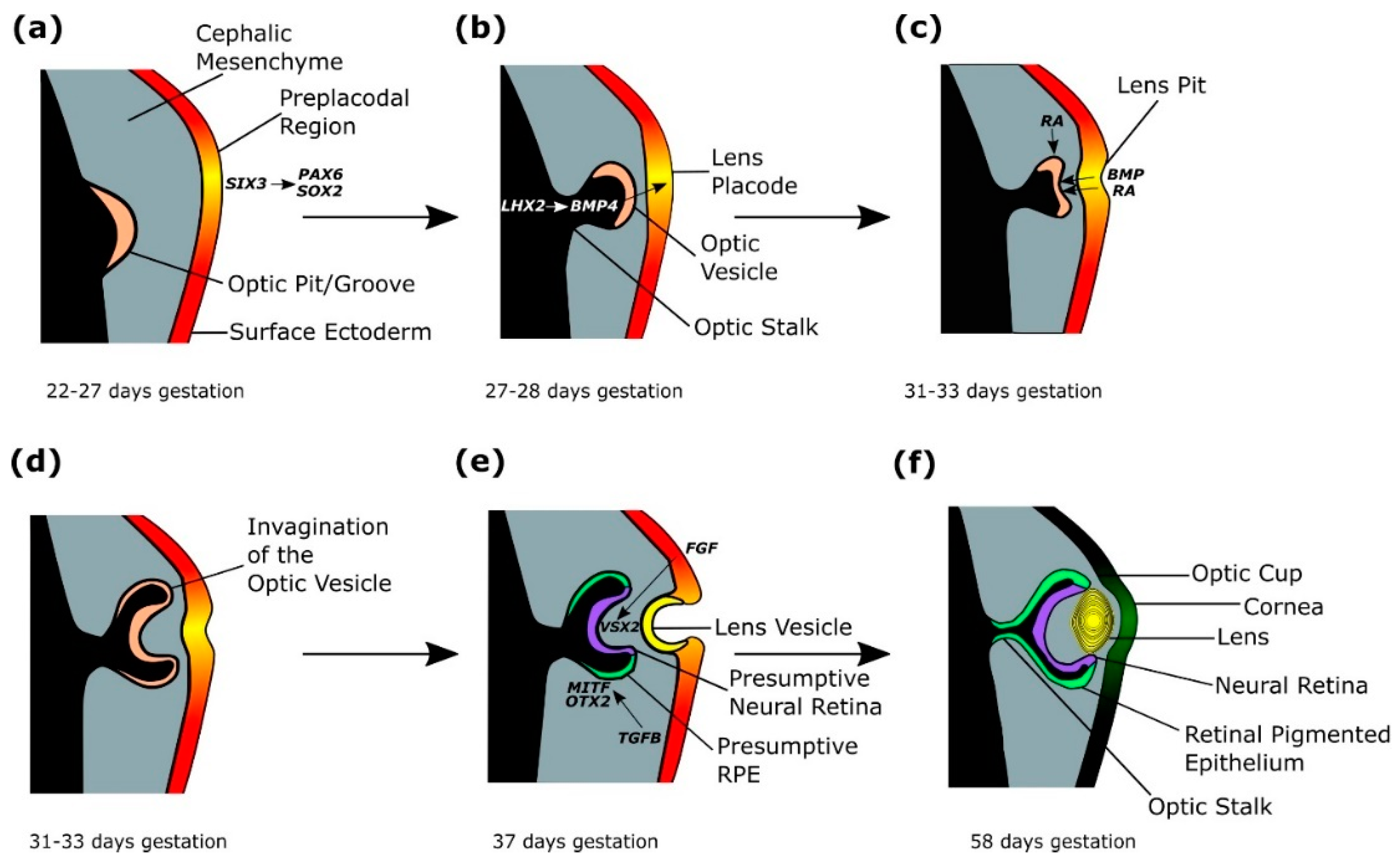
2.1. Eye Field Specification
2.2. Splitting of the Eye Field
2.3. Lens Placode Formation
2.4. Optic Cup Formation
3. The Genetic Basis of Anophthalmia and Microphthalmia
3.1. Eye Field Initiating Transcription Factors
3.1.1. SOX2
3.1.2. OTX2
3.1.3. RAX
3.1.4. VSX2
3.1.5. PAX6
3.2. Retinoic Acid Signalling Pathway Components
3.2.1. STRA6
3.2.2. RARβ
3.2.3. ALDH1A3
4. Current Research and Future Directions
4.1. Modelling Eye Development Using iPSCs
4.2. Genomics and Epigenetics
4.3. Proteomics and Metabolomics
4.4. Therapeutics
5. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Graw, J. Eye Development. Curr. Top. Dev. Biol. 2010, 90, 343–386. [Google Scholar] [CrossRef]
- Sinn, R. An eye on eye development. Mech. Dev. 2013, 130, 347–358. [Google Scholar] [CrossRef]
- Chow, R.L.; Lang, R.A. Early Eye Development in Vertebrates. Annu. Rev. Cell Dev. Biol. 2001, 17, 255–296. [Google Scholar] [CrossRef] [Green Version]
- Fuhrmann, S. Eye Morphogenesis and Patterning of the Optic Vesicle. Curr. Top. Dev. Biol. 2010, 93, 61–84. [Google Scholar] [CrossRef] [Green Version]
- FitzPatrick, D.R.; van Heyningen, V. Developmental eye disorders. Curr. Opin. Genet. Dev. 2005, 15, 348–353. [Google Scholar] [CrossRef]
- Plaisancié, J.; Ceroni, F.; Holt, R.; Zazo Seco, C.; Calvas, P.; Chassaing, N.; Ragge, N.K. Genetics of anophthalmia and microphthalmia. Part 1: Non-syndromic anophthalmia/microphthalmia. Hum. Genet. 2019, 1–32. [Google Scholar] [CrossRef]
- Chassaing, N.; Causse, A.; Vigouroux, A.; Delahaye, A.; Alessandri, J.-L.; Boespflug-Tanguy, O.; Boute-Benejean, O.; Dollfus, H.; Duban-Bedu, B.; Gilbert-Dussardier, B.; et al. Molecular findings and clinical data in a cohort of 150 patients with anophthalmia/microphthalmia. Clin. Genet. 2014, 86, 326–334. [Google Scholar] [CrossRef]
- Slavotinek, A. Genetics of anophthalmia and microphthalmia. Part 2: Syndromes associated with anophthalmia–microphthalmia. Hum. Genet. 2018, 1–16. [Google Scholar] [CrossRef]
- Verma, A.S.; FitzPatrick, D.R. Anophthalmia and microphthalmia. Orphanet J. Rare Dis. 2007, 2, 47. [Google Scholar] [CrossRef]
- Schneider, A.; Bardakjian, T.; Reis, L.M.; Tyler, R.C.; Semina, E.V. Novel SOX2 mutations and genotype-phenotype correlation in anophthalmia and microphthalmia. Am. J. Med. Genet. Part A 2009, 149A, 2706–2715. [Google Scholar] [CrossRef]
- Richardson, R.; Sowden, J.; Gerth-Kahlert, C.; Moore, A.T.; Moosajee, M. Clinical utility gene card for: Non-Syndromic Microphthalmia Including Next-Generation Sequencing-Based Approaches. Eur. J. Hum. Genet. 2017, 25. [Google Scholar] [CrossRef]
- Williamson, K.A.; FitzPatrick, D.R. The genetic architecture of microphthalmia, anophthalmia and coloboma. Eur. J. Med. Genet. 2014, 57, 369–380. [Google Scholar] [CrossRef]
- Shah, S.P.; Taylor, A.E.; Sowden, J.C.; Ragge, N.K.; Russell-Eggitt, I.; Rahi, J.S.; Gilbert, C.E. Anophthalmos, Microphthalmos, and Typical Coloboma in the United Kingdom: A Prospective Study of Incidence and Risk. Investig. Opthalmol. Vis. Sci. 2011, 52, 558–564. [Google Scholar] [CrossRef]
- Bernstein, C.S.; Anderson, M.T.; Gohel, C.; Slater, K.; Gross, J.M.; Agarwala, S. The cellular bases of choroid fissure formation and closure. Dev. Biol. 2018, 440, 137–151. [Google Scholar] [CrossRef]
- Gregory-Evans, C.Y.; Williams, M.J.; Halford, S.; Gregory-Evans, K. Ocular coloboma: A reassessment in the age of molecular neuroscience. J. Med. Genet. 2004, 41, 881–891. [Google Scholar] [CrossRef]
- Ragge, N.K.; Subak-Sharpe, I.D.; Collin, J.R.O. A practical guide to the management of anophthalmia and microphthalmia. Eye 2007, 21, 1290–1300. [Google Scholar] [CrossRef] [Green Version]
- Slavotinek, A.M. Eye development genes and known syndromes. Mol. Genet. Metab. 2011, 104, 448–456. [Google Scholar] [CrossRef] [Green Version]
- Wyatt, A.; Bakrania, P.; Bunyan, D.J.; Osborne, R.J.; Crolla, J.A.; Salt, A.; Ayuso, C.; Newbury-Ecob, R.; Abou-Rayyah, Y.; Collin, J.R.O.; et al. Novel heterozygous OTX2 mutations and whole gene deletions in anophthalmia, microphthalmia and coloboma. Hum. Mutat. 2008, 29, E278–E283. [Google Scholar] [CrossRef]
- Gerth-Kahlert, C.; Williamson, K.; Ansari, M.; Rainger, J.K.; Hingst, V.; Zimmermann, T.; Tech, S.; Guthoff, R.F.; van Heyningen, V.; FitzPatrick, D.R. Clinical and mutation analysis of 51 probands with anophthalmia and/or severe microphthalmia from a single center. Mol. Genet. Genomic Med. 2013, 1, 15–31. [Google Scholar] [CrossRef]
- Casey, J.; Kawaguchi, R.; Morrissey, M.; Sun, H.; McGettigan, P.; Nielsen, J.E.; Conroy, J.; Regan, R.; Kenny, E.; Cormican, P.; et al. First implication of STRA6 mutations in isolated anophthalmia, microphthalmia, and coloboma: A new dimension to the STRA6 phenotype. Hum. Mutat. 2011, 32, 1417–1426. [Google Scholar] [CrossRef]
- Slavotinek, A.M.; Garcia, S.T.; Chandratillake, G.; Bardakjian, T.; Ullah, E.; Wu, D.; Umeda, K.; Lao, R.; Ling, P.; Tang, P.L.-F.; et al. Exome Sequencing in 32 Patients with Anophthalmia/Microphthalmia and Developmental Eye Defects. Clin. Genet. 2015, 88, 468–473. [Google Scholar] [CrossRef]
- Givens, K.T.; Lee, D.A.; Jones, T.; Ilstrup, D.M. Congenital rubella syndrome: Ophthalmic manifestations and associated systemic disorders. Br. J. Ophthalmol. 1993, 77, 358–363. [Google Scholar] [CrossRef]
- Busby, A.; Dolk, H.; Armstrong, B. Eye anomalies: Seasonal variation and maternal viral infections. Epidemiology 2005, 16, 317–322. [Google Scholar] [CrossRef]
- Zagozewski, J.; Zhang, Q.; Eisenstat, D. Genetic regulation of vertebrate eye development. Clin. Genet. 2014, 86, 453–460. [Google Scholar] [CrossRef]
- Zuber, M.E.; Gestri, G.; Viczian, A.S.; Barsacchi, G.; Harris, W.A. Specification of the vertebrate eye by a network of eye field transcription factors. Development 2003, 130, 5155–5167. [Google Scholar] [CrossRef] [Green Version]
- Loosli, F.; Winkler, S.; Wittbrodt, J. Six3 overexpression initiates the formation of ectopic retina. Genes Dev. 1999, 13, 649–654. [Google Scholar] [CrossRef]
- Chuang, J.C.; Raymond, P.A. Embryonic origin of the eyes in teleost fish. BioEssays 2002, 24, 519–529. [Google Scholar] [CrossRef]
- Danno, H.; Michiue, T.; Hitachi, K.; Yukita, A.; Ishiura, S.; Asashima, M. Molecular links among the causative genes for ocular malformation: Otx2 and Sox2 coregulate Rax expression. Proc. Natl. Acad. Sci. USA 2008, 105, 5408–5413. [Google Scholar] [CrossRef]
- Medina-Martinez, O.; Amaya-Manzanares, F.; Liu, C.; Mendoza, M.; Shah, R.; Zhang, L.; Behringer, R.R.; Mahon, K.A.; Jamrich, M. Cell-Autonomous Requirement for Rx Function in the Mammalian Retina and Posterior Pituitary. PLoS ONE 2009, 4, e4513. [Google Scholar] [CrossRef]
- Mathers, P.H.; Grinberg, A.; Mahon, K.A.; Jamrich, M. The Rx homeobox gene is essential for vertebrate eye development. Nature 1997, 387, 603–607. [Google Scholar] [CrossRef]
- Lagutin, O.V.; Zhu, C.C.; Kobayashi, D.; Topczewski, J.; Shimamura, K.; Puelles, L.; Russell, H.R.C.; McKinnon, P.J.; Solnica-Krezel, L.; Oliver, G. Six3 repression of Wnt signaling in the anterior neuroectoderm is essential for vertebrate forebrain development. Genes Dev. 2003, 17, 368–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zagozewski, J.; Zhang, Q.; Pinto, V.; Wigle, J.; Eisenstat, D. The role of homeobox genes in retinal development and disease. Dev. Biol. 2014, 393, 195–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bazin-Lopez, N.; Valdivia, L.E.; Wilson, S.W.; Gestri, G. Watching eyes take shape. Curr. Opin. Genet. Dev. 2015, 32, 73–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiang, C.; Litingtung, Y.; Lee, E.; Young, K.E.; Corden, J.L.; Westphal, H.; Beachy, P.A. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature 1996, 383, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Geng, X.; Speirs, C.; Lagutin, O.; Inbal, A.; Liu, W.; Solnica-Krezel, L.; Jeong, Y.; Epstein, D.J.; Oliver, G. Haploinsufficiency of Six3 Fails to Activate Sonic hedgehog Expression in the Ventral Forebrain and Causes Holoprosencephaly. Dev. Cell 2008, 15, 236–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carl, M.; Wittbrodt, J. Graded interference with FGF signalling reveals its dorsoventral asymmetry at the mid-hindbrain boundary. Development 1999, 126, 5659–5667. [Google Scholar] [PubMed]
- Rebagliati, M.R.; Toyama, R.; Haffter, P.; Dawid, I.B. Cyclops encodes a nodal-related factor involved in midline signaling. Proc. Natl. Acad. Sci. USA 1998, 95, 9932–9937. [Google Scholar] [CrossRef]
- Varga, Z.M.; Wegner, J.; Westerfield, M. Anterior movement of ventral diencephalic precursors separates the primordial eye field in the neural plate and requires cyclops. Development 1999, 126, 5533–5546. [Google Scholar]
- Brown, K.E.; Keller, P.J.; Ramialison, M.; Rembold, M.; Stelzer, E.H.K.; Loosli, F.; Wittbrodt, J. Nlcam modulates midline convergence during anterior neural plate morphogenesis. Dev. Biol. 2010, 339, 14–25. [Google Scholar] [CrossRef] [Green Version]
- Rembold, M.; Loosli, F.; Adams, R.J.; Wittbrodt, J. Individual Cell Migration Serves as the Driving Force for Optic Vesicle Evagination. Science 2006, 313, 1130–1134. [Google Scholar] [CrossRef] [Green Version]
- Cvekl, A.; Ashery-Padan, R. The cellular and molecular mechanisms of vertebrate lens development. Development 2014, 141, 4432–4447. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Lagutin, O.V.; Mende, M.; Streit, A.; Oliver, G. Six3 activation of Pax6 expression is essential for mammalian lens induction and specification. EMBO J. 2006, 25, 5383–5395. [Google Scholar] [CrossRef] [PubMed]
- Cvekl, A.; McGreal, R.; Liu, W. Lens Development and Crystallin Gene Expression. Prog. Mol. Biol. Transl. Sci. 2015, 134, 129–167. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-T.; Kim, J.W. Compartmentalization of vertebrate optic neuroephithelium: External cues and transcription factors. Mol. Cells 2012, 33, 317–324. [Google Scholar] [CrossRef] [Green Version]
- Furuta, Y.; Hogan, B.L.M. BMP4 is essential for lens induction in the mouse embryo. Genes Dev. 1998, 12, 3764–3775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yun, S.; Saijoh, Y.; Hirokawa, K.E.; Kopinke, D.; Murtaugh, L.C.; Monuki, E.S.; Levine, E.M. Lhx2 links the intrinsic and extrinsic factors that control optic cup formation. Development 2009, 136, 3895–3906. [Google Scholar] [CrossRef] [PubMed]
- Hyer, J.; Kuhlman, J.; Afif, E.; Mikawa, T. Optic cup morphogenesis requires pre-lens ectoderm but not lens differentiation. Dev. Biol. 2003, 259, 351–363. [Google Scholar] [CrossRef]
- Cvekl, A.; Wang, W.-L. Retinoic acid signaling in mammalian eye development. Exp. Eye Res. 2009, 89, 280–291. [Google Scholar] [CrossRef] [Green Version]
- Mic, F.A.; Molotkov, A.; Molotkova, N.; Duester, G. Raldh2 expression in optic vesicle generates a retinoic acid signal needed for invagination of retina during optic cup formation. Dev. Dyn. 2004, 231, 270–277. [Google Scholar] [CrossRef]
- Molotkov, A.; Molotkova, N.; Duester, G.; Lemke, G.; Bertuzzi, S. Retinoic acid guides eye morphogenetic movements via paracrine signaling but is unnecessary for retinal dorsoventral patterning. Development 2006, 133, 1901–1910. [Google Scholar] [CrossRef] [Green Version]
- Matt, N.; Ghyselinck, N.B.; Pellerin, I.; Dupé, V. Impairing retinoic acid signalling in the neural crest cells is sufficient to alter entire eye morphogenesis. Dev. Biol. 2008, 320, 140–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dudley, A.T.; Robertson, E.J. Overlapping expression domains of bone morphogenetic protein family members potentially account for limited tissue defects in BMP7 deficient embryos. Dev. Dyn. 1997, 208, 349–362. [Google Scholar] [CrossRef]
- Chauhan, B.K.; Disanza, A.; Choi, S.-Y.; Faber, S.C.; Lou, M.; Beggs, H.E.; Scita, G.; Zheng, Y.; Lang, R.A. Cdc42-and IRSp53-dependent contractile filopodia tether presumptive lens and retina to coordinate epithelial invagination. Development 2009, 136, 3657–3667. [Google Scholar] [CrossRef] [PubMed]
- Oltean, A.; Huang, J.; Beebe, D.C.; Taber, L.A. Tissue growth constrained by extracellular matrix drives invagination during optic cup morphogenesis. Biomech. Model. Mechanobiol. 2016, 15, 1405–1421. [Google Scholar] [CrossRef] [PubMed]
- Plageman, T.F.; Chauhan, B.K.; Yang, C.; Jaudon, F.; Shang, X.; Zheng, Y.; Lou, M.; Debant, A.; Hildebrand, J.D.; Lang, R.A. A Trio-RhoA-Shroom3 pathway is required for apical constriction and epithelial invagination. Development 2011, 138, 5177–5188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; Rajagopal, R.; Liu, Y.; Dattilo, L.K.; Shaham, O.; Ashery-Padan, R.; Beebe, D.C. The mechanism of lens placode formation: A case of matrix-mediated morphogenesis. Dev. Biol. 2011, 355, 32–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plageman, T.F.; Chung, M.-I.; Lou, M.; Smith, A.N.; Hildebrand, J.D.; Wallingford, J.B.; Lang, R.A. Pax6-dependent Shroom3 expression regulates apical constriction during lens placode invagination. Development 2010, 137, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Fuhrmann, S.; Levine, E.M.; Reh, T.A. Extraocular mesenchyme patterns the optic vesicle during early eye development in the embryonic chick. Development 2000, 127, 4599–4609. [Google Scholar]
- Hägglund, A.-C.; Berghard, A.; Carlsson, L. Canonical Wnt/β-Catenin Signalling Is Essential for Optic Cup Formation. PLoS ONE 2013, 8, e81158. [Google Scholar] [CrossRef]
- Nguyen, M.; Arnheiter, H.; Fuhrmann, S. Signaling and transcriptional regulation in early mammalian eye development: A link between FGF and MITF. Development 2000, 127, 3581–3591. [Google Scholar]
- Capowski, E.E.; Simonett, J.M.; Clark, E.M.; Wright, L.S.; Howden, S.E.; Wallace, K.A.; Petelinsek, A.M.; Pinilla, I.; Phillips, M.J.; Meyer, J.S.; et al. Loss of MITF expression during human embryonic stem cell differentiation disrupts retinal pigment epithelium development and optic vesicle cell proliferation. Hum. Mol. Genet. 2014, 23, 6332–6344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-Morales, J.R.; Dolez, V.; Rodrigo, I.; Zaccarini, R.; Leconte, L.; Bovolenta, P.; Saule, S. OTX2 activates the molecular network underlying retina pigment epithelium differentiation. J. Biol. Chem. 2003, 278, 21721–21731. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Morales, J.R.; Signore, M.; Acampora, D.; Simeone, A.; Bovolenta, P. Otx genes are required for tissue specification in the developing eye. Development 2001, 128, 2019–2030. [Google Scholar] [PubMed]
- Vogel-Höpker, A.; Momose, T.; Rohrer, H.; Yasuda, K.; Ishihara, L.; Rapaport, D.H. Multiple functions of fibroblast growth factor-8 (FGF-8) in chick eye development. Mech. Dev. 2000, 94, 25–36. [Google Scholar] [CrossRef]
- Liang, L.; Sandell, J.H. Focus on molecules: Homeobox protein Chx10. Exp. Eye Res. 2008, 86, 541–542. [Google Scholar] [CrossRef] [PubMed]
- Burkitt Wright, E.M.M.; Perveen, R.; Bowers, N.; Ramsden, S.; McCann, E.; O’Driscoll, M.; Lloyd, I.C.; Clayton-Smith, J.; Black, G.C.M. VSX2 in microphthalmia: A novel splice site mutation producing a severe microphthalmia phenotype. Br. J. Ophthalmol. 2010, 94, 386–388. [Google Scholar] [CrossRef] [PubMed]
- Wong, G.; Conger, S.B.; Burmeister, M. Mapping of genetic modifiers affecting the eye phenotype of ocular retardation (Chx10or-J) mice. Mamm. Genome 2006, 17, 518–525. [Google Scholar] [CrossRef]
- Horsford, D.J.; Nguyen, M.-T.T.; Sellar, G.C.; Kothary, R.; Arnheiter, H.; McInnes, R.R. Chx10 repression of Mitf is required for the maintenance of mammalian neuroretinal identity. Development 2005, 132, 177–187. [Google Scholar] [CrossRef]
- Burmeister, M.; Novak, J.; Liang, M.-Y.; Basu, S.; Ploder, L.; Hawes, N.L.; Vidgen, D.; Hoover, F.; Goldman, D.; Kalnins, V.I.; et al. Ocular retardation mouse caused by Chx10 homeobox null allele: Impaired retinal progenitor proliferation and bipolar cell differentiation. Nat. Genet. 1996, 12, 376–384. [Google Scholar] [CrossRef]
- Marquardt, T.; Ashery-Padan, R.; Andrejewski, N.; Scardigli, R.; Guillemot, F.; Gruss, P. Pax6 Is Required for the Multipotent State of Retinal Progenitor Cells. Cell 2001, 105, 43–55. [Google Scholar] [CrossRef] [Green Version]
- Torres, M.; Gómez-Pardo, E.; Gruss, P. Pax2 contributes to inner ear patterning and optic nerve trajectory. Development 1996, 122, 3381–3391. [Google Scholar] [PubMed]
- Schwarz, M.; Cecconi, F.; Bernier, G.; Andrejewski, N.; Kammandel, B.; Wagner, M.; Gruss, P. Spatial specification of mammalian eye territories by reciprocal transcriptional repression of Pax2 and Pax6. Development 2000, 127, 4325–4334. [Google Scholar]
- Hallonet, M.; Hollemann, T.; Pieler, T.; Gruss, P. Vax1, a novel homeobox-containing gene, directs development of the basal forebrain and visual system. Genes Dev. 1999, 13, 3106–3114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Take-uchi, M.; Clarke, J.D.W.; Wilson, S.W. Hedgehog signalling maintains the optic stalk-retinal interface through the regulation of Vax gene activity. Development 2003, 130, 955–968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mui, S.H.; Kim, J.W.; Lemke, G.; Bertuzzi, S. Vax genes ventralize the embryonic eye. Genes Dev. 2005, 19, 1249–1259. [Google Scholar] [CrossRef]
- Richardson, R.; Tracey-White, D.; Webster, A.; Moosajee, M. The zebrafish eye-a paradigm for investigating human ocular genetics. Eye 2017, 31, 68–86. [Google Scholar] [CrossRef]
- Liu, C.; Nathans, J.; Bovolenta, P.; Alfano, G.; Marchitiello, A.; Mocchetti, C.; Crippa, L.; Bulfone, A.; Marigo, V.; Ballabio, A.; et al. An essential role for frizzled 5 in mammalian ocular development. Development 2008, 135, 3567–3576. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Bakeri, H.; Li, T.; Swaroop, A. Regulation of retinal progenitor expansion by Frizzled receptors: Implications for microphthalmia and retinal coloboma. Hum. Mol. Genet. 2012, 21, 1848–1860. [Google Scholar] [CrossRef]
- Knickmeyer, M.D.; Mateo, J.L.; Eckert, P.; Roussa, E.; Rahhal, B.; Zuniga, A.; Krieglstein, K.; Wittbrodt, J.; Heermann, S. TGFβ-facilitated optic fissure fusion and the role of bone morphogenetic protein antagonism. Open Biol. 2018, 8, 170134. [Google Scholar] [CrossRef]
- Ragge, N.K.; Brown, A.G.; Poloschek, C.M.; Lorenz, B.; Henderson, R.A.; Clarke, M.P.; Russell-Eggitt, I.; Fielder, A.; Gerrelli, D.; Martinez-Barbera, J.P.; et al. Heterozygous Mutations of OTX2 Cause Severe Ocular Malformations. Am. J. Hum. Genet. 2005, 76, 1008–1022. [Google Scholar] [CrossRef] [Green Version]
- Riera, M.; Wert, A.; Nieto, I.; Pomares, E. Panel-based whole exome sequencing identifies novel mutations in microphthalmia and anophthalmia patients showing complex Mendelian inheritance patterns. Mol. Genet. Genomic Med. 2017, 5, 709–719. [Google Scholar] [CrossRef] [PubMed]
- Biesecker, L.G.; Spinner, N.B. A genomic view of mosaicism and human disease. Nat. Rev. Genet. 2013, 14, 307–320. [Google Scholar] [CrossRef] [PubMed]
- Chassaing, N.; Gilbert-Dussardier, B.; Nicot, F.; Fermeaux, V.; Encha-Razavi, F.; Fiorenza, M.; Toutain, A.; Calvas, P. Germinal mosaicism and familial recurrence of aSOX2 mutation with highly variable phenotypic expression extending from AEG syndrome to absence of ocular involvement. Am. J. Med. Genet. Part A 2007, 143A, 289–291. [Google Scholar] [CrossRef] [PubMed]
- Faivre, L.; Williamson, K.A.; Faber, V.; Laurent, N.; Grimaldi, M.; Thauvin-Robinet, C.; Durand, C.; Mugneret, F.; Gouyon, J.-B.; Bron, A.; et al. Recurrence of SOX2 anophthalmia syndrome with gonosomal mosaicism in a phenotypically normal mother. Am. J. Med. Genet. Part A 2006, 140A, 636–639. [Google Scholar] [CrossRef] [PubMed]
- Plaisancie, J.; Calvas, P.; Chassaing, N. Genetic Advances in Microphthalmia. J. Pediatr. Genet. 2016, 5, 184–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, S.P.; Taylor, A.E.; Sowden, J.C.; Ragge, N.; Russell-Eggitt, I.; Rahi, J.S.; Gilbert, C.E. Anophthalmos, Microphthalmos, and Coloboma in the United Kingdom: Clinical Features, Results of Investigations, and Early Management. Ophthalmology 2012, 119, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Srour, M.; Chitayat, D.; Caron, V.; Chassaing, N.; Bitoun, P.; Patry, L.; Cordier, M.-P.; Capo-Chichi, J.-M.; Francannet, C.; Calvas, P.; et al. Recessive and Dominant Mutations in Retinoic Acid Receptor Beta in Cases with Microphthalmia and Diaphragmatic Hernia. Am. J. Hum. Genet. 2013, 93, 765–772. [Google Scholar] [CrossRef] [Green Version]
- Srour, M.; Caron, V.; Pearson, T.; Nielsen, S.B.; Lévesque, S.; Delrue, M.-A.; Becker, T.A.; Hamdan, F.F.; Kibar, Z.; Sattler, S.G.; et al. Gain-of-Function Mutations in RARB Cause Intellectual Disability with Progressive Motor Impairment. Hum. Mutat. 2016, 37, 786–793. [Google Scholar] [CrossRef]
- Ramirez-Botero, A.F.; Pachajoa, H. Syndromic microphthalmia-3 caused by a mutation on gene SOX2 in a Colombian male patient. Congenit. Anom. 2016, 56, 250–252. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Rodriguez, J.; Pelcastre, E.L.; Tovilla-Canales, J.L.; Garcia-Ortiz, J.E.; Amato-Almanza, M.; Villanueva-Mendoza, C.; Espinosa-Mattar, Z.; Zenteno, J.C. Mutational screening of CHX10, GDF6, OTX2, RAX and SOX2 genes in 50 unrelated microphthalmia-anophthalmia-coloboma (MAC) spectrum cases. Br. J. Ophthalmol. 2010, 94, 1100–1104. [Google Scholar] [CrossRef]
- Schilter, K.F.; Schneider, A.; Bardakjian, T.; Soucy, J.-F.; Tyler, R.C.; Reis, L.M.; Semina, E.V. OTX2 microphthalmia syndrome: Four novel mutations and delineation of a phenotype. Clin. Genet. 2011, 79, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Deml, B.; Reis, L.M.; Lemyre, E.; Clark, R.D.; Kariminejad, A.; Semina, E.V. Novel mutations in PAX6, OTX2 and NDP in anophthalmia, microphthalmia and coloboma. Eur. J. Hum. Genet. 2016, 24, 535–541. [Google Scholar] [CrossRef] [PubMed]
- Tajima, T.; Ohtake, A.; Hoshino, M.; Amemiya, S.; Sasaki, N.; Ishizu, K.; Fujieda, K. OTX2 Loss of Function Mutation Causes Anophthalmia and Combined Pituitary Hormone Deficiency with a Small Anterior and Ectopic Posterior Pituitary. J. Clin. Endocrinol. Metab. 2009, 94, 314–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voronina, V.A.; Kozhemyakina, E.A.; O’Kernick, C.M.; Kahn, N.D.; Wenger, S.L.; Linberg, J.V.; Schneider, A.S.; Mathers, P.H. Mutations in the human RAX homeobox gene in a patient with anophthalmia and sclerocornea. Hum. Mol. Genet. 2003, 13, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Iseri, S.U.; Wyatt, A.W.; Nürnberg, G.; Kluck, C.; Nürnberg, P.; Holder, G.E.; Blair, E.; Salt, A.; Ragge, N.K. Use of genome-wide SNP homozygosity mapping in small pedigrees to identify new mutations in VSX2 causing recessive microphthalmia and a semidominant inner retinal dystrophy. Hum. Genet. 2010, 128, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Chassaing, N.; Ragge, N.; Kariminejad, A.; Buffet, A.; Ghaderi-Sohi, S.; Martinovic, J.; Calvas, P. Mutation analysis of the STRA6 gene in isolated and non-isolated anophthalmia/microphthalmia. Clin. Genet. 2013, 83, 244–250. [Google Scholar] [CrossRef]
- Pasutto, F.; Sticht, H.; Hammersen, G.; Gillessen-Kaesbach, G.; FitzPatrick, D.R.; Nürnberg, G.; Brasch, F.; Schirmer-Zimmermann, H.; Tolmie, J.L.; Chitayat, D.; et al. Mutations in STRA6 Cause a Broad Spectrum of Malformations Including Anophthalmia, Congenital Heart Defects, Diaphragmatic Hernia, Alveolar Capillary Dysplasia, Lung Hypoplasia, and Mental Retardation. Am. J. Hum. Genet. 2007, 80, 550–560. [Google Scholar] [CrossRef] [Green Version]
- Chassaing, N.; Golzio, C.; Odent, S.; Lequeux, L.; Vigouroux, A.; Martinovic-Bouriel, J.; Tiziano, F.D.; Masini, L.; Piro, F.; Maragliano, G.; et al. Phenotypic spectrum of STRA6 mutations: From Matthew-Wood syndrome to non-lethal anophthalmia. Hum. Mutat. 2009, 30, E673–E681. [Google Scholar] [CrossRef]
- Fares-Taie, L.; Gerber, S.; Chassaing, N.; Clayton-Smith, J.; Hanein, S.; Silva, E.; Serey, M.; Serre, V.; Gérard, X.; Baumann, C.; et al. ALDH1A3 Mutations Cause Recessive Anophthalmia and Microphthalmia. Am. J. Hum. Genet. 2013, 92, 265–270. [Google Scholar] [CrossRef] [Green Version]
- Abouzeid, H.; Favez, T.; Schmid, A.; Agosti, C.; Youssef, M.; Marzouk, I.; El Shakankiry, N.; Bayoumi, N.; Munier, F.L.; Schorderet, D.F. Mutations in ALDH1A3 Represent a Frequent Cause of Microphthalmia/Anophthalmia in Consanguineous Families. Hum. Mutat. 2014, 35, 949–953. [Google Scholar] [CrossRef]
- Plaisancié, J.; Brémond-Gignac, D.; Demeer, B.; Gaston, V.; Verloes, A.; Fares-Taie, L.; Gerber, S.; Rozet, J.-M.; Calvas, P.; Chassaing, N. Incomplete penetrance of biallelic ALDH1A3 mutations. Eur. J. Med. Genet. 2016, 59, 215–218. [Google Scholar] [CrossRef] [PubMed]
- Bakrania, P.; Efthymiou, M.; Klein, J.C.; Salt, A.; Bunyan, D.J.; Wyatt, A.; Ponting, C.P.; Martin, A.; Williams, S.; Lindley, V.; et al. Mutations in BMP4 Cause Eye, Brain, and Digit Developmental Anomalies: Overlap between the BMP4 and Hedgehog Signaling Pathways. Am. J. Hum. Genet. 2008, 82, 304–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reis, L.M.; Tyler, R.C.; Schilter, K.F.; Abdul-Rahman, O.; Innis, J.W.; Kozel, B.A.; Schneider, A.S.; Bardakjian, T.M.; Lose, E.J.; Martin, D.M.; et al. BMP4 loss-of-function mutations in developmental eye disorders including SHORT syndrome. Hum. Genet. 2011, 130, 495–504. [Google Scholar] [CrossRef] [Green Version]
- Wyatt, A.W.; Osborne, R.J.; Stewart, H.; Ragge, N.K. Bone morphogenetic protein 7 (BMP7) mutations are associated with variable ocular, brain, ear, palate, and skeletal anomalies. Hum. Mutat. 2010, 31, 781–787. [Google Scholar] [CrossRef] [PubMed]
- Ye, M.; Berry-Wynne, K.M.; Asai-Coakwell, M.; Sundaresan, P.; Footz, T.; French, C.R.; Abitbol, M.; Fleisch, V.C.; Corbett, N.; Allison, W.T.; et al. Mutation of the bone morphogenetic protein GDF3 causes ocular and skeletal anomalies. Hum. Mol. Genet. 2010, 19, 287–298. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; He, F.; Bu, J.; Liu, X.; Du, W.; Dong, J.; Cooney, J.D.; Dubey, S.K.; Shi, Y.; Gong, B.; et al. ABCB6 Mutations Cause Ocular Coloboma. Am. J. Hum. Genet. 2012, 90, 40–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, K.; Logan, C.V.; McKibbin, M.; Sheridan, E.; Elçioglu, N.H.; Yenice, O.; Parry, D.A.; Fernandez-Fuentes, N.; Abdelhamed, Z.I.A.; Al-Maskari, A.; et al. Next generation sequencing identifies mutations in Atonal homolog 7 (ATOH7) in families with global eye developmental defects. Hum. Mol. Genet. 2012, 21, 776–783. [Google Scholar] [CrossRef]
- Zahrani, F.; Aldahmesh, M.A.; Alshammari, M.J.; Al-Hazzaa, S.A.F.; Alkuraya, F.S. Mutations in C12orf57 Cause a Syndromic Form of Colobomatous Microphthalmia. Am. J. Hum. Genet. 2013, 92, 387–391. [Google Scholar] [CrossRef] [Green Version]
- Chassaing, N.; Ragge, N.; Plaisancié, J.; Patat, O.; Geneviève, D.; Rivier, F.; Malrieu-Eliaou, C.; Hamel, C.; Kaplan, J.; Calvas, P. Confirmation of TENM3 involvement in autosomal recessive colobomatous microphthalmia. Am. J. Med. Genet. Part A 2016, 170, 1895–1898. [Google Scholar] [CrossRef]
- Aldahmesh, M.A.; Mohammed, J.Y.; Al-Hazzaa, S.; Alkuraya, F.S. Homozygous null mutation in ODZ3 causes microphthalmia in humans. Genet. Med. 2012, 14, 900–904. [Google Scholar] [CrossRef] [Green Version]
- Slavotinek, A.M.; Chao, R.; Vacik, T.; Yahyavi, M.; Abouzeid, H.; Bardakjian, T.; Schneider, A.; Shaw, G.; Sherr, E.H.; Lemke, G.; et al. VAX1 mutation associated with microphthalmia, corpus callosum agenesis, and orofacial clefting: The first description of a VAX1 phenotype in humans. Hum. Mutat. 2012, 33, 364–368. [Google Scholar] [CrossRef] [PubMed]
- Okada, I.; Hamanoue, H.; Terada, K.; Tohma, T.; Megarbane, A.; Chouery, E.; Abou-Ghoch, J.; Jalkh, N.; Cogulu, O.; Ozkinay, F.; et al. SMOC1 Is Essential for Ocular and Limb Development in Humans and Mice. Am. J. Hum. Genet. 2011, 88, 30–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kondo, Y.; Koshimizu, E.; Megarbane, A.; Hamanoue, H.; Okada, I.; Nishiyama, K.; Kodera, H.; Miyatake, S.; Tsurusaki, Y.; Nakashima, M.; et al. Whole-exome sequencing identified a homozygous FNBP4 mutation in a family with a condition similar to microphthalmia with limb anomalies. Am. J. Med. Genet. Part A 2013, 161, 1543–1546. [Google Scholar] [CrossRef] [PubMed]
- Schimmenti, L.A.; de la Cruz, J.; Lewis, R.A.; Karkera, J.D.; Manligas, G.S.; Roessler, E.; Muenke, M. Novel mutation in sonic hedgehog in non-syndromic colobomatous microphthalmia. Am. J. Med. Genet. 2003, 116, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Bakrania, P.; Ugur Iseri, S.A.; Wyatt, A.W.; Bunyan, D.J.; Lam, W.W.K.; Salt, A.; Ramsay, J.; Robinson, D.O.; Ragge, N.K. Sonic hedgehog mutations are an uncommon cause of developmental eye anomalies. Am. J. Med. Genet. Part A 2010, 152A, 1310–1313. [Google Scholar] [CrossRef] [PubMed]
- Esmailpour, T.; Riazifar, H.; Liu, L.; Donkervoort, S.; Huang, V.H.; Madaan, S.; Shoucri, B.M.; Busch, A.; Wu, J.; Towbin, A.; et al. A splice donor mutation in NAA10 results in the dysregulation of the retinoic acid signalling pathway and causes Lenz microphthalmia syndrome. J. Med. Genet. 2014, 51, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Ragge, N.; Isidor, B.; Bitoun, P.; Odent, S.; Giurgea, I.; Cogné, B.; Deb, W.; Vincent, M.; Le Gall, J.; Morton, J.; et al. Expanding the phenotype of the X-linked BCOR microphthalmia syndromes. Hum. Genet. 2018, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Wimplinger, I.; Morleo, M.; Rosenberger, G.; Iaconis, D.; Orth, U.; Meinecke, P.; Lerer, I.; Ballabio, A.; Gal, A.; Franco, B.; et al. Mutations of the Mitochondrial Holocytochrome c–Type Synthase in X-Linked Dominant Microphthalmia with Linear Skin Defects Syndrome. Am. J. Hum. Genet. 2006, 79, 878–889. [Google Scholar] [CrossRef]
- Rainger, J.; Pehlivan, D.; Johansson, S.; Bengani, H.; Sanchez-Pulido, L.; Williamson, K.A.; Ture, M.; Barker, H.; Rosendahl, K.; Spranger, J.; et al. Monoallelic and Biallelic Mutations in MAB21L2 Cause a Spectrum of Major Eye Malformations. Am. J. Hum. Genet. 2014, 94, 915–923. [Google Scholar] [CrossRef] [Green Version]
- Chou, C.M.; Nelson, C.; Tarlé, S.A.; Pribila, J.T.; Bardakjian, T.; Woods, S.; Schneider, A.; Glaser, T. Biochemical Basis for Dominant Inheritance, Variable Penetrance, and Maternal Effects in RBP4 Congenital Eye Disease. Cell 2015, 161, 634–646. [Google Scholar] [CrossRef] [Green Version]
- Bertolacini, C.D.P.; Ribeiro-Bicudo, L.A.; Petrin, A.; Richieri-Costa, A.; Murray, J.C. Clinical findings in patients with GLI2 mutations-phenotypic variability. Clin. Genet. 2012, 81, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Guion-Almeida, M.L.; Richieri-Costa, A.; Zechi-Ceide, R.M. Holoprosencephaly spectrum, ano/microphthalmia, and first branchial arch defects: Evidence for a new disorder. Clin. Dysmorphol. 2008, 17, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, L.A.; Guerini Rde, C.; Richieri-Costa, A. Holoprosencephaly with microphthalmia, hypoplastic ears, vertebral segmentation defects, and congenital heart defects. Am. J. Med. Genet. Part A 2005, 136A, 350–351. [Google Scholar] [CrossRef] [PubMed]
- Froyen, G.; Govaerts, K.; Van Esch, H.; Verbeeck, J.; Tuomi, M.-L.; HeikkilÃ, H.; Torniainen, S.; Devriendt, K.; Fryns, J.-P.; Marynen, P.; et al. Novel PORCN mutations in focal dermal hypoplasia. Clin. Genet. 2009, 76, 535–543. [Google Scholar] [CrossRef]
- Leoyklang, P.; Suphapeetiporn, K.; Wananukul, S.; Shotelersuk, V. Three novel mutations in the PORCN gene underlying focal dermal hypoplasia. Clin. Genet. 2008, 73, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Slavotinek, A.; Li, C.; Sherr, E.H.; Chudley, A.E. Mutation analysis of the FRAS1 gene demonstrates new mutations in a propositus with Fraser syndrome. Am. J. Med. Genet. Part A 2006, 140A, 1909–1914. [Google Scholar] [CrossRef] [PubMed]
- Slavotinek, A.M.; Baranzini, S.E.; Schanze, D.; Labelle-Dumais, C.; Short, K.M.; Chao, R.; Yahyavi, M.; Bijlsma, E.K.; Chu, C.; Musone, S.; et al. Manitoba-oculo-tricho-anal (MOTA) syndrome is caused by mutations in FREM1. J. Med. Genet. 2011, 48, 375–382. [Google Scholar] [CrossRef]
- Shaw, N.D.; Brand, H.; Kupchinsky, Z.A.; Bengani, H.; Plummer, L.; Jones, T.I.; Erdin, S.; Williamson, K.A.; Rainger, J.; Stortchevoi, A.; et al. SMCHD1 mutations associated with a rare muscular dystrophy can also cause isolated arhinia and Bosma arhinia microphthalmia syndrome. Nat. Genet. 2017, 49, 238–248. [Google Scholar] [CrossRef] [Green Version]
- Gallardo, M.E.; Rodríguez de Córdoba, S.; Schneider, A.S.; Dwyer, M.A.; Ayuso, C.; Bovolenta, P. Analysis of the developmental SIX6 homeobox gene in patients with anophthalmia/microphthalmia. Am. J. Med. Genet. Part A 2004, 129, 92–94. [Google Scholar] [CrossRef]
- Gestri, G.; Osborne, R.J.; Wyatt, A.W.; Gerrelli, D.; Gribble, S.; Stewart, H.; Fryer, A.; Bunyan, D.J.; Prescott, K.; Collin, J.R.O.; et al. Reduced TFAP2A function causes variable optic fissure closure and retinal defects and sensitizes eye development to mutations in other morphogenetic regulators. Hum. Genet. 2009, 126, 791–803. [Google Scholar] [CrossRef] [Green Version]
- Shaheen, R.; Faqeih, E.; Seidahmed, M.Z.; Sunker, A.; Alali, F.E.; Khadijah, A.; Alkuraya, F.S. A TCTN2 mutation defines a novel Meckel Gruber syndrome locus. Hum. Mutat. 2011, 32, 573–578. [Google Scholar] [CrossRef] [PubMed]
- Shaheen, R.; Shamseldin, H.E.; Loucks, C.M.; Seidahmed, M.Z.; Ansari, S.; Ibrahim Khalil, M.; Al-Yacoub, N.; Davis, E.E.; Mola, N.A.; Szymanska, K.; et al. Mutations in CSPP1, Encoding a Core Centrosomal Protein, Cause a Range of Ciliopathy Phenotypes in Humans. Am. J. Hum. Genet. 2014, 94, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Deml, B.; Reis, L.M.; Maheshwari, M.; Griffis, C.; Bick, D.; Semina, E.V. Whole exome analysis identifies dominant COL4A1 mutations in patients with complex ocular phenotypes involving microphthalmia. Clin. Genet. 2014, 86, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.; Ellard, S.; Kneen, R.; Lim, M.; Osborne, N.; Rankin, J.; Stoodley, N.; Van Der Knaap, M.; Whitney, A.; Jardine, P. Childhood presentation of COL4A1 mutations. Dev. Med. Child Neurol. 2012, 54, 569–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chassaing, N.; Davis, E.E.; McKnight, K.L.; Niederriter, A.R.; Causse, A.; David, V.; Desmaison, A.; Lamarre, S.; Vincent-Delorme, C.; Pasquier, L.; et al. Targeted resequencing identifies PTCH1 as a major contributor to ocular developmental anomalies and extends the SOX2 regulatory network. Genome Res. 2016, 26, 474–485. [Google Scholar] [CrossRef]
- Adly, N.; Alhashem, A.; Ammari, A.; Alkuraya, F.S. Ciliary Genes TBC1D32/C6orf170 and SCLT1 are Mutated in Patients with OFD Type IX. Hum. Mutat. 2014, 35, 36–40. [Google Scholar] [CrossRef]
- Matías-Pérez, D.; García-Montaño, L.A.; Cruz-Aguilar, M.; García-Montalvo, I.A.; Nava-Valdéz, J.; Barragán-Arevalo, T.; Villanueva-Mendoza, C.; Villarroel, C.E.; Guadarrama-Vallejo, C.; la Cruz, R.V.; et al. Identification of novel pathogenic variants and novel gene-phenotype correlations in Mexican subjects with microphthalmia and/or anophthalmia by next-generation sequencing. J. Hum. Genet. 2018, 63, 1169–1180. [Google Scholar] [CrossRef]
- Aramaki, M.; Udaka, T.; Kosaki, R.; Makita, Y.; Okamoto, N.; Yoshihashi, H.; Oki, H.; Nanao, K.; Moriyama, N.; Oku, S.; et al. Phenotypic spectrum of CHARGE syndrome with CHD7 mutations. J. Pediatr. 2006, 148, 410–414. [Google Scholar] [CrossRef]
- Nishina, S.; Kosaki, R.; Yagihashi, T.; Azuma, N.; Okamoto, N.; Hatsukawa, Y.; Kurosawa, K.; Yamane, T.; Mizuno, S.; Tsuzuki, K.; et al. Ophthalmic features of CHARGE syndrome with CHD7 mutations. Am. J. Med. Genet. Part A 2012, 158A, 514–518. [Google Scholar] [CrossRef]
- Crespí, J.; Buil, J.A.; Bassaganyas, F.; Vela-Segarra, J.I.; Díaz-Cascajosa, J.; Ayala-Ramírez, R.; Zenteno, J.C. A Novel Mutation Confirms MFRP as the Gene Causing the Syndrome of Nanophthalmos–Renititis Pigmentosa–Foveoschisis–Optic Disk Drusen. Am. J. Ophthalmol. 2008, 146, 323–328. [Google Scholar] [CrossRef]
- Nair, K.S.; Hmani-Aifa, M.; Ali, Z.; Kearney, A.L.; Ben Salem, S.; Macalinao, D.G.; Cosma, I.M.; Bouassida, W.; Hakim, B.; Benzina, Z.; et al. Alteration of the serine protease PRSS56 causes angle-closure glaucoma in mice and posterior microphthalmia in humans and mice. Nat. Genet. 2011, 43, 579–584. [Google Scholar] [CrossRef] [PubMed]
- Gal, A.; Rau, I.; El Matri, L.; Kreienkamp, H.-J.; Fehr, S.; Baklouti, K.; Chouchane, I.; Li, Y.; Rehbein, M.; Fuchs, J.; et al. Autosomal-Recessive Posterior Microphthalmos Is Caused by Mutations in PRSS56, a Gene Encoding a Trypsin-Like Serine Protease. Am. J. Hum. Genet. 2011, 88, 382–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khorram, D.; Choi, M.; Roos, B.R.; Stone, E.M.; Kopel, T.; Allen, R.; Alward, W.L.M.; Scheetz, T.E.; Fingert, J.H. Novel TMEM98 mutations in pedigrees with autosomal dominant nanophthalmos. Mol. Vis. 2015, 21, 1017–1023. [Google Scholar] [PubMed]
- Awadalla, M.S.; Burdon, K.P.; Souzeau, E.; Landers, J.; Hewitt, A.W.; Sharma, S.; Craig, J.E. Mutation in TMEM98 in a Large White Kindred With Autosomal Dominant Nanophthalmos Linked to 17p12-q12. JAMA Ophthalmol. 2014, 132, 970–977. [Google Scholar] [CrossRef] [PubMed]
- Scott, A.F.; Mohr, D.W.; Kasch, L.M.; Barton, J.A.; Pittiglio, R.; Ingersoll, R.; Craig, B.; Marosy, B.A.; Doheny, K.F.; Bromley, W.C.; et al. Identification of an HMGB3 Frameshift Mutation in a Family With an X-linked Colobomatous Microphthalmia Syndrome Using Whole-Genome and X-Exome Sequencing. JAMA Ophthalmol. 2014, 132, 1215–1220. [Google Scholar] [CrossRef] [PubMed]
- Choi, A.; Lao, R.; Ling-Fung Tang, P.; Wan, E.; Mayer, W.; Bardakjian, T.; Shaw, G.M.; Kwok, P.; Schneider, A.; Slavotinek, A. Novel mutations in PXDN cause microphthalmia and anterior segment dysgenesis. Eur. J. Hum. Genet. 2015, 23, 337–341. [Google Scholar] [CrossRef]
- Chao, R.; Nevin, L.; Agarwal, P.; Riemer, J.; Bai, X.; Delaney, A.; Akana, M.; JimenezLopez, N.; Bardakjian, T.; Schneider, A.; et al. A Male with Unilateral Microphthalmia Reveals a Role for TMX3 in Eye Development. PLoS ONE 2010, 5, e10565. [Google Scholar] [CrossRef]
- Williamson, K.A.; Rainger, J.; Floyd, J.A.B.; Ansari, M.; Meynert, A.; Aldridge, K.V.; Rainger, J.K.; Anderson, C.A.; Moore, A.T.; Hurles, M.E.; et al. Heterozygous Loss-of-Function Mutations in YAP1 Cause Both Isolated and Syndromic Optic Fissure Closure Defects. Am. J. Hum. Genet. 2014, 94, 295–302. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.-F.; Xiang, L.; Cheng, W.; Cheng, F.-F.; He, K.-W.; Zhang, B.-W.; Zheng, S.-S.; Han, R.-Y.; Zheng, Y.-H.; Xu, X.-T.; et al. Mutation of IPO13 causes recessive ocular coloboma, microphthalmia, and cataract. Exp. Mol. Med. 2018, 50, 53. [Google Scholar] [CrossRef]
- Bidinost, C.; Matsumoto, M.; Chung, D.; Salem, N.; Zhang, K.; Stockton, D.W.; Khoury, A.; Megarbane, A.; Bejjani, B.A.; Traboulsi, E.I. Heterozygous and Homozygous Mutations in PITX3 in a Large Lebanese Family with Posterior Polar Cataracts and Neurodevelopmental Abnormalities. Investig. Opthalmol. Vis. Sci. 2006, 47, 1274–1280. [Google Scholar] [CrossRef]
- George, A.; Zand, D.J.; Hufnagel, R.B.; Sharma, R.; Sergeev, Y.V.; Legare, J.M.; Rice, G.M.; Scott Schwoerer, J.A.; Rius, M.; Tetri, L.; et al. Biallelic Mutations in MITF Cause Coloboma, Osteopetrosis, Microphthalmia, Macrocephaly, Albinism, and Deafness. Am. J. Hum. Genet. 2016, 99, 1388–1394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaur, K.; Ragge, N.K.; Ragoussis, J. Molecular analysis of FOXC1 in subjects presenting with severe developmental eye anomalies. Mol. Vis. 2009, 15, 1366–1373. [Google Scholar] [PubMed]
- Willer, T.; Lee, H.; Lommel, M.; Yoshida-Moriguchi, T.; de Bernabe, D.B.V.; Venzke, D.; Cirak, S.; Schachter, H.; Vajsar, J.; Voit, T.; et al. ISPD loss-of-function mutations disrupt dystroglycan O-mannosylation and cause Walker-Warburg syndrome. Nat. Genet. 2012, 44, 575–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vetro, A.; Iascone, M.; Limongelli, I.; Ameziane, N.; Gana, S.; Della Mina, E.; Giussani, U.; Ciccone, R.; Forlino, A.; Pezzoli, L.; et al. Loss-of-Function FANCL Mutations Associate with Severe Fanconi Anemia Overlapping the VACTERL Association. Hum. Mutat. 2015, 36, 562–568. [Google Scholar] [CrossRef] [PubMed]
- Twigg, S.R.F.; Hufnagel, R.B.; Miller, K.A.; Zhou, Y.; McGowan, S.J.; Taylor, J.; Craft, J.; Taylor, J.C.; Santoro, S.L.; Huang, T.; et al. A Recurrent Mosaic Mutation in SMO, Encoding the Hedgehog Signal Transducer Smoothened, Is the Major Cause of Curry-Jones Syndrome. Am. J. Hum. Genet. 2016, 98, 1256–1265. [Google Scholar] [CrossRef] [PubMed]
- Sukalo, M.; Tilsen, F.; Kayserili, H.; Müller, D.; Tüysüz, B.; Ruddy, D.M.; Wakeling, E.; Ørstavik, K.H.; Snape, K.M.; Trembath, R.; et al. DOCK6 Mutations Are Responsible for a Distinct Autosomal-Recessive Variant of Adams-Oliver Syndrome Associated with Brain and Eye Anomalies. Hum. Mutat. 2015, 36, 593–598. [Google Scholar] [CrossRef]
- Song, Z.; Si, N.; Xiao, W. A novel mutation in the CRYAA gene associated with congenital cataract and microphthalmia in a Chinese family. BMC Med. Genet. 2018, 19, 190. [Google Scholar] [CrossRef]
- Asgari, N.; Akbari, M.T.; Deilamani, F.K.; Babamohammadi, G. Molecular Genetic Analysis of FOXL2 Gene in Two Iranian Families with Blepharophimosis-Ptosis-Epicanthus Inversus Syndrome. J. Hum. Genet. Genomics 2017, 1. [Google Scholar] [CrossRef]
- Billingsley, G.; Santhiya, S.T.; Paterson, A.D.; Ogata, K.; Wodak, S.; Hosseini, S.M.; Manisastry, S.M.; Vijayalakshmi, P.; Gopinath, P.M.; Graw, J.; et al. CRYBA4, a Novel Human Cataract Gene, Is Also Involved in Microphthalmia. Am. J. Hum. Genet. 2006, 79, 702–709. [Google Scholar] [CrossRef] [Green Version]
- Kou, Y.; Shboul, M.; Wang, Z.; Shersheer, Q.; Lyu, Z.; Liu, P.; Zhao, X.; Tian, J. Novel frame shift mutation in ERCC6 leads to a severe form of Cockayne syndrome with postnatal growth failure and early death: A case report and brief literature review. Medicine 2018, 97, e11636. [Google Scholar] [CrossRef]
- Laugel, V.; Dalloz, C.; Tobias, E.S.; Tolmie, J.L.; Martin-Coignard, D.; Drouin-Garraud, V.; Valayannopoulos, V.; Sarasin, A.; Dollfus, H. Cerebro-oculo-facio-skeletal syndrome: Three additional cases with CSB mutations, new diagnostic criteria and an approach to investigation. J. Med. Genet. 2008, 45, 564–571. [Google Scholar] [CrossRef] [PubMed]
- Drury, S.; Boustred, C.; Tekman, M.; Stanescu, H.; Kleta, R.; Lench, N.; Chitty, L.S.; Scott, R.H. A Novel Homozygous ERCC5 Truncating Mutation in a Family With Prenatal Arthrogryposis-Further Evidence of Genotype-Phenotype Correlation. Am. J. Med. Genet. Part A 2014, 164, 1777–1783. [Google Scholar] [CrossRef] [PubMed]
- Jaspers, N.G.J.; Raams, A.; Silengo, M.C.; Wijgers, N.; Niedernhofer, L.J.; Robinson, A.R.; Giglia-Mari, G.; Hoogstraten, D.; Kleijer, W.J.; Hoeijmakers, J.H.J.; et al. First Reported Patient with Human ERCC1 Deficiency Has Cerebro-Oculo-Facio-Skeletal Syndrome with a Mild Defect in Nucleotide Excision Repair and Severe Developmental Failure. Am. J. Hum. Genet. 2007, 80, 457–466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cantagrel, V.; Lefeber, D.J.; Ng, B.G.; Guan, Z.; Silhavy, J.L.; Bielas, S.L.; Lehle, L.; Hombauer, H.; Adamowicz, M.; Swiezewska, E.; et al. SRD5A3 Is Required for Converting Polyprenol to Dolichol and Is Mutated in a Congenital Glycosylation Disorder. Cell 2010, 142, 203–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ullah, E.; Wu, D.; Madireddy, L.; Lao, R.; Ling-Fung Tang, P.; Wan, E.; Bardakjian, T.; Kopinsky, S.; Kwok, P.-Y.; Schneider, A.; et al. Two missense mutations in SALL4 in a patient with microphthalmia, coloboma, and optic nerve hypoplasia. Ophthalmic Genet. 2017, 38, 371–375. [Google Scholar] [CrossRef]
- De Bernardo, G.; Giordano, M.; Di Toro, A.; Sordino, D.; De Brasi, D. Prenatal diagnosis of Fraser syndrome: A matter of life or death? Ital. J. Pediatr. 2015, 41, 86. [Google Scholar] [CrossRef]
- Devuyst, O.; Arnould, V.J. Mutations in RPGRIP1L: Extending the clinical spectrum of ciliopathies. Nephrol. Dial. Transplant. 2008, 23, 1500–1503. [Google Scholar] [CrossRef]
- Ehmke, N.; Graul-Neumann, L.; Smorag, L.; Koenig, R.; Segebrecht, L.; Magoulas, P.; Scaglia, F.; Kilic, E.; Hennig, A.F.; Adolphs, N.; et al. De Novo Mutations in SLC25A24 Cause a Craniosynostosis Syndrome with Hypertrichosis, Progeroid Appearance, and Mitochondrial Dysfunction. Am. J. Hum. Genet. 2017, 101, 833–843. [Google Scholar] [CrossRef] [Green Version]
- Unger, S.; Górna, M.W.; Le Béchec, A.; Do Vale-Pereira, S.; Bedeschi, M.F.; Geiberger, S.; Grigelioniene, G.; Horemuzova, E.; Lalatta, F.; Lausch, E.; et al. FAM111A Mutations Result in Hypoparathyroidism and Impaired Skeletal Development. Am. J. Hum. Genet. 2013, 92, 990–995. [Google Scholar] [CrossRef] [Green Version]
- Shaheen, R.; Anazi, S.; Ben-Omran, T.; Seidahmed, M.Z.; Caddle, L.B.; Palmer, K.; Ali, R.; Alshidi, T.; Hagos, S.; Goodwin, L.; et al. Mutations in SMG9, Encoding an Essential Component of Nonsense-Mediated Decay Machinery, Cause a Multiple Congenital Anomaly Syndrome in Humans and Mice. Am. J. Hum. Genet. 2016, 98, 643–652. [Google Scholar] [CrossRef] [Green Version]
- Wallis, D.E.; Roessler, E.; Hehr, U.; Nanni, L.; Wiltshire, T.; Richieri-Costa, A.; Gillessen-Kaesbach, G.; Zackai, E.H.; Rommens, J.; Muenke, M. Mutations in the homeodomain of the human SIX3 gene cause holoprosencephaly. Nat. Genet. 1999, 22, 196–198. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.; Wright, K.J.; Le Corre, S.; Micalizzi, A.; Romani, M.; Abhyankar, A.; Saada, J.; Perrault, I.; Amiel, J.; Litzler, J.; et al. A Homozygous PDE6D Mutation in Joubert Syndrome Impairs Targeting of Farnesylated INPP5E Protein to the Primary Cilium. Hum. Mutat. 2014, 35, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Bögershausen, N.; Altunoglu, U.; Beleggia, F.; Yigit, G.; Kayserili, H.; Nürnberg, P.; Li, Y.; Altmüller, J.; Wollnik, B. An unusual presentation of Kabuki syndrome with orbital cysts, microphthalmia, and cholestasis with bile duct paucity. Am. J. Med. Genet. Part A 2016, 170, 3282–3288. [Google Scholar] [CrossRef] [PubMed]
- Amiel, J.; Audollent, S.; Joly, D.; Dureau, P.; Salomon, R.; Tellier, A.-L.; Augé, J.; Bouissou, F.; Antignac, C.; Gubler, M.-C.; et al. PAX2 mutations in renal-coloboma syndrome: Mutational hotspot and germline mosaicism. Eur. J. Hum. Genet. 2000, 8, 820–826. [Google Scholar] [CrossRef] [PubMed]
- Bower, M.; Salomon, R.; Allanson, J.; Antignac, C.; Benedicenti, F.; Benetti, E.; Binenbaum, G.; Jensen, U.B.; Cochat, P.; DeCramer, S.; et al. Update of PAX2 mutations in renal coloboma syndrome and establishment of a locus-specific database. Hum. Mutat. 2012, 33, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Valente, E.M.; Logan, C.V.; Mougou-Zerelli, S.; Lee, J.H.; Silhavy, J.L.; Brancati, F.; Iannicelli, M.; Travaglini, L.; Romani, S.; Illi, B.; et al. Mutations in TMEM216 perturb ciliogenesis and cause Joubert, Meckel and related syndromes. Nat. Genet. 2010, 42, 619–625. [Google Scholar] [CrossRef] [PubMed]
- Baala, L.; Audollent, S.; Martinovic, J.; Ozilou, C.; Babron, M.-C.; Sivanandamoorthy, S.; Saunier, S.; Salomon, R.; Gonzales, M.; Rattenberry, E.; et al. Pleiotropic Effects of CEP290 (NPHP6) Mutations Extend to Meckel Syndrome. Am. J. Hum. Genet. 2007, 81, 170–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, G.E.; Ostergaard, P.; Moore, A.T.; Connell, F.C.; Williams, D.; Quarrell, O.; Brady, A.F.; Spier, I.; Hazan, F.; Moldovan, O.; et al. Microcephaly with or without chorioretinopathy, lymphoedema, or mental retardation (MCLMR): Review of phenotype associated with KIF11 mutations. Eur. J. Hum. Genet. 2014, 22, 881–887. [Google Scholar] [CrossRef]
- Li, J.-K.; Fei, P.; Li, Y.; Huang, Q.-J.; Zhang, Q.; Zhang, X.; Rao, Y.-Q.; Li, J.; Zhao, P. Identification of novel KIF11 mutations in patients with familial exudative vitreoretinopathy and a phenotypic analysis. Sci. Rep. 2016, 6, 26564. [Google Scholar] [CrossRef] [Green Version]
- Vervoort, V.S.; Viljoen, D.; Smart, R.; Suthers, G.; DuPont, B.R.; Abbott, A.; Schwartz, C.E. Sorting nexin 3 (SNX3) is disrupted in a patient with a translocation t(6;13) (q21;q12) and microcephaly, microphthalmia, ectrodactyly, prognathism (MMEP) phenotype. J. Med. Genet. 2002, 39, 893–899. [Google Scholar] [CrossRef]
- Ariss, M.; Natan, K.; Friedman, N.; Traboulsi, E.I. Ophthalmologic Abnormalities in Mowat-Wilson Syndrome and a Mutation in ZEB2. Ophthalmic Genet. 2012, 33, 159–160. [Google Scholar] [CrossRef] [PubMed]
- Beltrán-Valero De Bernabé, D.; Currier, S.; Steinbrecher, A.; Celli, J.; Van Beusekom, E.; Van Der Zwaag, B.; Lya Kayserili, H.; Merlini, L.; Chitayat, D.; Dobyns, W.B.; et al. Mutations in the O-Mannosyltransferase Gene POMT1 Give Rise to the Severe Neuronal Migration Disorder Walker-Warburg Syndrome. Am. J. Hum. Genet 2002, 71, 1033–1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.S.; Hayashi, Y.K.; Matsumoto, H.; Ogawa, M.; Noguchi, S.; Murakami, N.; Sakuta, R.; Mochizuki, M.; Michele, D.E.; Campbell, K.P.; et al. POMT1 mutation results in defective glycosylation and loss of laminin-binding activity in alpha-DG. Neurology 2004, 62, 1009–1011. [Google Scholar] [CrossRef] [PubMed]
- Van Reeuwijk, J.; Janssen, M.; Van Den Elzen, C.; Beltran-Valero De Bernabé, D.; Sabatelli, P.; Merlini, L.; Boon, M.; Scheffer, H.; Brockington, M.; Muntoni, F.; et al. POMT2 mutations cause a-dystroglycan hypoglycosylation and Walker-Warburg syndrome. J. Med. Genet. 2005, 42, 907–912. [Google Scholar] [CrossRef] [PubMed]
- Teber, S.; Sezer, T.; Kafalı, M.; Chiara Manzini, M.; Konuk Yüksel, B.; Tekin, M.; Fitöz, S.; Walsh, C.A.; Deda, G. Severe muscle–eye–brain disease is associated with a homozygous mutation in the POMGnT1 gene. Eur. J. Paediatr. Neurol. 2008, 12, 133–136. [Google Scholar] [CrossRef]
- Kondo-Iida, E.; Kobayashi, K.; Watanabe, M.; Sasaki, J.; Kumagai, T.; Koide, H.; Saito, K.; Osawa, M.; Nakamura, Y.; Toda, T. Novel Mutations and Genotype-Phenotype Relationships in 107 Families With Fukuyama-Type Congenital Muscular Dystrophy (FCMD). Hum. Mol. Genet. 1999, 8, 2303–2309. [Google Scholar] [CrossRef] [PubMed]
- Beltran-Valero De Bernabé, D.; Voit, T.; Longman, C.; Steinbrecher, A.; Straub, V.; Yuva, Y.; Herrmann, R.; Sperner, J.; Korenke, C.; Diesen, C.; et al. Mutations in the FKRP gene can cause muscle-eye-brain disease and Walker-Warburg syndrome. J. Med. Genet. 2004, 41, 61. [Google Scholar] [CrossRef]
- Van Reeuwijk, J.; Olderode-Berends, M.J.W.; Van Den Elzen, C.; Brouwer, O.F.; Roscioli, T.; Van Pampus, M.G.; Scheffer, H.; Brunner, H.G.; Van Bokhoven, H.; Hol, F.A. A homozygous FKRP start codon mutation is associated with Walker-Warburg syndrome, the severe end of the clinical spectrum. Clin. Genet. 2010, 78, 275–281. [Google Scholar] [CrossRef] [Green Version]
- Leibovitz, Z.; Mandel, H.; Falik-Zaccai, T.C.; Ben Harouch, S.; Savitzki, D.; Krajden-Haratz, K.; Gindes, L.; Tamarkin, M.; Lev, D.; Dobyns, W.B.; et al. Walker-Warburg syndrome and tectocerebellar dysraphia: A novel association caused by a homozygous DAG1 mutation. Eur. J. Paediatr. Neurol. 2018, 22, 525–531. [Google Scholar] [CrossRef]
- Riemersma, M.; Mandel, H.; van Beusekom, E.; Gazzoli, I.; Roscioli, T.; Eran, A.; Gershoni-Baruch, R.; Gershoni, M.; Pietrokovski, S.; Vissers, L.E.; et al. Absence of α- and β-dystroglycan is associated with Walker-Warburg syndrome. Neurology 2015, 84, 2177–2182. [Google Scholar] [CrossRef]
- Stevens, E.; Carss, K.J.; Cirak, S.; Foley, A.R.; Torelli, S.; Willer, T.; Tambunan, D.E.; Yau, S.; Brodd, L.; Sewry, C.A.; et al. Mutations in B3GALNT2 Cause Congenital Muscular Dystrophy and Hypoglycosylation of a-Dystroglycan. Am. J. Hum. Genet. 2013, 92, 354–365. [Google Scholar] [CrossRef] [PubMed]
- Handley, M.T.; Morris-Rosendahl, D.J.; Brown, S.; Macdonald, F.; Hardy, C.; Bem, D.; Carpanini, S.M.; Borck, G.; Martorell, L.; Izzi, C.; et al. Mutation Spectrum in RAB3GAP1, RAB3GAP2, and RAB18 and Genotype-Phenotype Correlations in Warburg Micro Syndrome and Martsolf Syndrome. Hum. Mutat. 2013, 34, 686–696. [Google Scholar] [CrossRef] [PubMed]
- Borck, G.; Wunram, H.; Steiert, A.; Volk, A.E.; Körber, F.; Roters, S.; Herkenrath, P.; Wollnik, B.; Morris-Rosendahl, D.J.; Kubisch, C. A homozygous RAB3GAP2 mutation causes Warburg Micro syndrome. Hum. Genet. 2011, 129, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Aligianis, I.A.; Morgan, N.V.; Mione, M.; Johnson, C.A.; Rosser, E.; Hennekam, R.C.; Adams, G.; Trembath, R.C.; Pilz, D.T.; Stoodley, N.; et al. Mutation in Rab3 GTPase-Activating Protein (RAB3GAP) Noncatalytic Subunit in a Kindred with Martsolf Syndrome. Am. J. Hum. Genet. 2006, 78, 702–707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Esch, H.; Jansen, A.; Bauters, M.; Froyen, G.; Fryns, J.-P. Encephalopathy and bilateral cataract in a boy with an interstitial deletion of Xp22 comprising the CDKL5 and NHS genes. Am. J. Med. Genet. Part A 2007, 143A, 364–369. [Google Scholar] [CrossRef]
- Gillespie, R.L.; Urquhart, J.; Lovell, S.C.; Biswas, S.; Parry, N.R.A.; Schorderet, D.F.; Lloyd, I.C.; Clayton-Smith, J.; Black, G.C. Abrogation of HMX1 Function Causes Rare Oculoauricular Syndrome Associated With Congenital Cataract, Anterior Segment Dysgenesis, and Retinal Dystrophy. Investig. Ophthalmol. Vis. Sci. 2015, 56, 883–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schorderet, D.F.; Nichini, O.; Boisset, G.; Polok, B.; Tiab, L.; Mayeur, H.; Raji, B.; de la Houssaye, G.; Abitbol, M.M.; Munier, F.L. Mutation in the Human Homeobox Gene NKX5-3 Causes an Oculo-Auricular Syndrome. Am. J. Hum. Genet. 2008, 82, 1178–1184. [Google Scholar] [CrossRef] [Green Version]
- Vitiello, C.; D’Adamo, P.; Gentile, F.; Vingolo, E.M.; Gasparini, P.; Banfi, S. A novel GJA1 mutation causes oculodentodigital dysplasia without syndactyly. Am. J. Med. Genet. Part A 2005, 133A, 58–60. [Google Scholar] [CrossRef]
- Richardson, R.J.; Joss, S.; Tomkin, S.; Ahmed, M.; Sheridan, E.; Dixon, M.J. A nonsense mutation in the first transmembrane domain of connexin 43 underlies autosomal recessive oculodentodigital syndrome. J. Med. Genet. 2006, 43, e37. [Google Scholar] [CrossRef]
- Brice, G.; Ostergaard, P.; Jeffery, S.; Gordon, K.; Mortimer, P.S.; Mansour, S. A novel mutation in GJA1 causing oculodentodigital syndrome and primary lymphoedema in a three generation family. Clin. Genet. 2013, 84, 378–381. [Google Scholar] [CrossRef]
- Narumi, S.; Numakura, C.; Shiihara, T.; Seiwa, C.; Nozaki, Y.; Yamagata, T.; Momoi, M.Y.; Watanabe, Y.; Yoshino, M.; Matsuishi, T.; et al. Various types of LRP5 mutations in four patients with osteoporosis-pseudoglioma syndrome: Identification of a 7.2-kb microdeletion using oligonucleotide tiling microarray. Am. J. Med. Genet. Part A 2010, 152A, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Garay, I.; Tomás, M.; Oltra, S.; Ramser, J.; Moltó, M.D.; Prieto, F.; Meindl, A.; Kutsche, K.; Martínez, F. A two base pair deletion in the PQBP1 gene is associated with microphthalmia, microcephaly, and mental retardation. Eur. J. Hum. Genet. 2007, 15, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Isrie, M.; Breuss, M.; Tian, G.; Hansen, A.H.; Cristofoli, F.; Morandell, J.; Kupchinsky, Z.A.; Sifrim, A.; Rodriguez-Rodriguez, C.M.; Dapena, E.P.; et al. Mutations in Either TUBB or MAPRE2 Cause Circumferential Skin Creases Kunze Type. Am. J. Hum. Genet. 2015, 97, 790–800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breuss, M.; Heng, J.I.-T.; Poirier, K.; Tian, G.; Jaglin, X.H.; Qu, Z.; Braun, A.; Gstrein, T.; Ngo, L.; Haas, M.; et al. Mutations in the β-Tubulin Gene TUBB5 Cause Microcephaly with Structural Brain Abnormalities. Cell Rep. 2012, 2, 1554–1562. [Google Scholar] [CrossRef] [PubMed]
- Bardakjian, T.M.; Schneider, A.S.; Ng, D.; Johnston, J.J.; Biesecker, L.G. Association of a de novo16q copy number variant with a phenotype that overlaps with Lenz microphthalmia and Townes-Brocks syndromes. BMC Med. Genet. 2009, 10, 137. [Google Scholar] [CrossRef] [PubMed]
- Chassaing, N.; Siani, V.; Carles, D.; Delezoide, A.L.; Alberti, E.M.; Battin, J.; Chateil, J.F.; Gilbert-Dussardier, B.; Coupry, I.; Arveiler, B.; et al. X-linked dominant chondrodysplasia with platyspondyly, distinctive brachydactyly, hydrocephaly, and microphthalmia. Am. J. Med. Genet. Part A 2005, 136A, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Simon, D.; Laloo, B.; Barillot, M.; Barnetche, T.; Blanchard, C.; Rooryck, C.; Marche, M.; Burgelin, I.; Coupry, I.; Chassaing, N.; et al. A mutation in the 3′-UTR of the HDAC6 gene abolishing the post-transcriptional regulation mediated by hsa-miR-433 is linked to a new form of dominant X-linked chondrodysplasia. Hum. Mol. Genet. 2010, 19, 2015–2027. [Google Scholar] [CrossRef]
- Uz, E.; Alanay, Y.; Aktas, D.; Vargel, I.; Gucer, S.; Tuncbilek, G.; von Eggeling, F.; Yilmaz, E.; Deren, O.; Posorski, N.; et al. Disruption of ALX1 Causes Extreme Microphthalmia and Severe Facial Clefting: Expanding the Spectrum of Autosomal-Recessive ALX-Related Frontonasal Dysplasia. Am. J. Hum. Genet. 2010, 86, 789–796. [Google Scholar] [CrossRef] [Green Version]
- Fregeau, B.; Kim, B.J.; Hernández-García, A.; Jordan, V.K.; Cho, M.T.; Schnur, R.E.; Monaghan, K.G.; Juusola, J.; Rosenfeld, J.A.; Bhoj, E.; et al. De Novo Mutations of RERE Cause a Genetic Syndrome with Features that Overlap Those Associated with Proximal 1p36 Deletions. Am. J. Hum. Genet. 2016, 98, 963–970. [Google Scholar] [CrossRef] [Green Version]
- Bem, D.; Yoshimura, S.-I.; Nunes-Bastos, R.; Bond, F.F.; Kurian, M.A.; Rahman, F.; Handley, M.T.W.; Hadzhiev, Y.; Masood, I.; Straatman-Iwanowska, A.A.; et al. Loss-of-Function Mutations in RAB18 Cause Warburg Micro Syndrome. Am. J. Hum. Genet. 2011, 88, 499–507. [Google Scholar] [CrossRef] [Green Version]
- Hingorani, M.; Hanson, I.; van Heyningen, V. Aniridia. Eur. J. Hum. Genet. 2012, 20, 1011–1017. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.; Thomas, M.G.; Andrews, C.; Chan, W.-M.; Proudlock, F.A.; McLean, R.J.; Pradeep, A.; Engle, E.C.; Gottlob, I. Autosomal-dominant nystagmus, foveal hypoplasia and presenile cataract associated with a novel PAX6 mutation. Eur. J. Hum. Genet. 2014, 22, 344–349. [Google Scholar] [CrossRef] [PubMed]
- Hall, H.N.; Williamson, K.A.; Hanson, I.M.; Owen, L.J.; Bengani, H.; Van, V.; Marsh, J.A.; Fitzpatrick, D.R. Recurrent heterozygous missense variants in the PAX6 paired domain cause severe microphthalmia. In Proceedings of the Association for Research in Vision and Ophthalmology (ARVO)-Poster Presented at Gene Variants and Regulation of Ocular Genes Expression in Health and Disease Session, Vancouver, BC, Canada, 28 April–2 May 2019; p. A0373. [Google Scholar]
- Taranova, O.V.; Magness, S.T.; Fagan, B.M.; Wu, Y.; Surzenko, N.; Hutton, S.R.; Pevny, L.H. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev. 2006, 20, 1187–1202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tajima, T.; Ishizu, K.; Nakamura, A. Molecular and Clinical Findings in Patients with LHX4 and OTX2 Mutations. Clin. Pediatr. Endocrinol. Case Reports Clin. Investig. Off. J. Jpn. Soc. Pediatr. Endocrinol. 2013, 22, 15–23. [Google Scholar] [CrossRef] [Green Version]
- Acampora, D.; Mazan, S.; Lallemand, Y.; Avantaggiato, V.; Maury, M.; Simeone, A.; Brulet, P. Forebrain and midbrain regions are deleted in Otx2−/− mutants due to a defective anterior neuroectoderm specification during gastrulation. Development 1995, 121, 3279–3290. [Google Scholar] [PubMed]
- Ang, S.L.; Jin, O.; Rhinn, M.; Daigle, N.; Stevenson, L.; Rossant, J. A targeted mouse Otx2 mutation leads to severe defects in gastrulation and formation of axial mesoderm and to deletion of rostral brain. Development 1996, 122, 243–252. [Google Scholar] [PubMed]
- Matsuo, I.; Kuratani, S.; Kimura, C.; Takeda, N.; Aizawa, S. Mouse Otx2 functions in the formation and patterning of rostral head. Genes Dev. 1995, 9, 2646–2658. [Google Scholar] [CrossRef]
- Hide, T.; Hatakeyama, J.; Kimura-Yoshida, C.; Tian, E.; Takeda, N.; Ushio, Y.; Shiroishi, T.; Aizawa, S.; Matsuo, I. Genetic modifiers of otocephalic phenotypes in Otx2 heterozygous mutant mice. Development 2002, 129, 4347–4357. [Google Scholar]
- Muranishi, Y.; Terada, K.; Inoue, T.; Katoh, K.; Tsujii, T.; Sanuki, R.; Kurokawa, D.; Aizawa, S.; Tamaki, Y.; Furukawa, T. An essential role for RAX homeoprotein and NOTCH-HES signaling in Otx2 expression in embryonic retinal photoreceptor cell fate determination. J. Neurosci. 2011, 31, 16792–16807. [Google Scholar] [CrossRef]
- Lupo, G.; Andreazzoli, M.; Gestri, G.; Liu, Y.; He, R.Q.; Barsacchi, G. Homeobox genes in the genetic control of eye development. Int. J. Dev. Biol. 2000, 44, 627–636. [Google Scholar]
- Andreazzoli, M.; Gestri, G.; Angeloni, D.; Menna, E.; Barsacchi, G. Role of Xrx1 in Xenopus eye and anterior brain development. Development 1999, 126, 2451–2460. [Google Scholar] [PubMed]
- Chow, R.L.; Altmann, C.R.; Lang, R.A.; Hemmati-Brivanlou, A. Pax6 induces ectopic eyes in a vertebrate. Development 1999, 126, 4213–4222. [Google Scholar] [PubMed]
- Nishihara, D.; Yajima, I.; Tabata, H.; Nakai, M.; Tsukiji, N.; Katahira, T.; Takeda, K.; Shibahara, S.; Nakamura, H.; Yamamoto, H. Otx2 Is Involved in the Regional Specification of the Developing Retinal Pigment Epithelium by Preventing the Expression of Sox2 and Fgf8, Factors That Induce Neural Retina Differentiation. PLoS ONE 2012, 7, e48879. [Google Scholar] [CrossRef] [PubMed]
- Tucker, P.; Laemle, L.; Munson, A.; Kanekar, S.; Oliver, E.R.; Brown, N.; Schlecht, H.; Vetter, M.; Glaser, T. The eyeless mouse mutation (ey1) removes an alternative start codon from theRx/rax homeobox gene. Genesis 2001, 31, 43–53. [Google Scholar] [CrossRef]
- Loosli, F.; Staub, W.; Finger-Baier, K.C.; Ober, E.A.; Verkade, H.; Wittbrodt, J.; Baier, H. Loss of eyes in zebrafish caused by mutation of chokh/rx3. EMBO Rep. 2003, 4, 894–899. [Google Scholar] [CrossRef]
- Muranishi, Y.; Terada, K.; Furukawa, T. An essential role for Rax in retina and neuroendocrine system development. Dev. Growth Differ. 2012, 54, 341–348. [Google Scholar] [CrossRef]
- Zhang, L.; Mathers, P.H.; Jamrich, M. Function of Rx, but not Pax6, is essential for the formation of retinal progenitor cells in mice. Genesis 2000, 28, 135–142. [Google Scholar] [CrossRef]
- Barabino, S.M.L.; Spada, F.; Cotelli, F.; Boncinelli, E. Inactivation of the zebrafish homologue of Chx10 by antisense oligonucleotides causes eye malformations similar to the ocular retardation phenotype. Mech. Dev. 1997, 63, 133–143. [Google Scholar] [CrossRef]
- Rowan, S.; Chen, C.-M.A.; Young, T.L.; Fisher, D.E.; Cepko, C.L.; McInnes, R.R. Transdifferentiation of the retina into pigmented cells in ocular retardation mice defines a new function of the homeodomain gene Chx10. Development 2004, 131, 5139–5152. [Google Scholar] [CrossRef]
- Green, E.S.; Stubbs, J.L.; Levine, E.M. Genetic rescue of cell number in a mouse model of microphthalmia: Interactions between Chx10 and G1-phase cell cycle regulators. Development 2003, 130, 539–552. [Google Scholar] [CrossRef]
- Capowski, E.E.; Wright, L.S.; Liang, K.; Phillips, M.J.; Wallace, K.; Petelinsek, A.; Hagstrom, A.; Pinilla, I.; Borys, K.; Lien, J.; et al. Regulation of WNT Signaling by VSX2 During Optic Vesicle Patterning in Human Induced Pluripotent Stem Cells. Stem Cells 2016, 34, 2625–2634. [Google Scholar] [CrossRef] [PubMed]
- Moosajee, M.; Hingorani, M.; Moore, A.T. PAX6-Related Aniridia. In GeneReviews®; University of Washington: Seattle, WA, USA, 2018. [Google Scholar]
- Kamachi, Y.; Uchikawa, M.; Tanouchi, A.; Sekido, R.; Kondoh, H. Pax6 and SOX2 form a co-DNA-binding partner complex that regulates initiation of lens development. Genes Dev. 2001, 15, 1272–1286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaham, O.; Menuchin, Y.; Farhy, C.; Ashery-Padan, R. Pax6: A multi-level regulator of ocular development. Prog. Retin. Eye Res. 2012, 31, 351–376. [Google Scholar] [CrossRef] [PubMed]
- Grindley, J.C.; Davidson, D.R.; Hill, R.E. The role of Pax-6 in eye and nasal development. Development 1995, 121, 1433–1442. [Google Scholar] [PubMed]
- Hill, R.E.; Favor, J.; Hogan, B.L.M.; Ton, C.C.T.; Saunders, G.F.; Hanson, I.M.; Prosser, J.; Jordan, T.; Hastie, N.D.; Heyningen, V. Van Mouse Small eye results from mutations in a paired-like homeobox-containing gene. Nature 1991, 354, 522–525. [Google Scholar] [CrossRef]
- Tétreault, N.; Champagne, M.-P.; Bernier, G. The LIM homeobox transcription factor Lhx2 is required to specify the retina field and synergistically cooperates with Pax6 for Six6 trans-activation. Dev. Biol. 2009, 327, 541–550. [Google Scholar] [CrossRef] [Green Version]
- Wawersik, S.; Purcell, P.; Rauchman, M.; Dudley, A.T.; Robertson, E.J.; Maas, R. BMP7 Acts in Murine Lens Placode Development. Dev. Biol. 1999, 207, 176–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, Z.; Tao, C.; Li, H.; Ladher, R.; Gotoh, N.; Feng, G.-S.; Wang, F.; Zhang, X.; Marigo, V.; Ballabio, A.; et al. Deficient FGF signaling causes optic nerve dysgenesis and ocular coloboma. Development 2013, 140, 2711–2723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segel, R.; Levy-Lahad, E.; Pasutto, F.; Picard, E.; Rauch, A.; Alterescu, G.; Schimmel, M.S. Pulmonary hypoplasia-diaphragmatic hernia-anophthalmia-cardiac defect (PDAC) syndrome due to STRA6 mutations-What are the minimal criteria? Am. J. Med. Genet. Part A 2009, 149A, 2457–2463. [Google Scholar] [CrossRef]
- Chitayat, D.; Sroka, H.; Keating, S.; Colby, R.S.; Ryan, G.; Toi, A.; Blaser, S.; Viero, S.; Devisme, L.; Boute-Bénéjean, O.; et al. The PDAC syndrome (pulmonary hypoplasia/agenesis, diaphragmatic hernia/eventration, anophthalmia/microphthalmia, and cardiac defect) (Spear syndrome, Matthew-Wood syndrome): Report of eight cases including a living child and further evidence for autosomal. Am. J. Med. Genet. Part A 2007, 143A, 1268–1281. [Google Scholar] [CrossRef]
- Kawaguchi, R.; Yu, J.; Honda, J.; Hu, J.; Whitelegge, J.; Ping, P.; Wiita, P.; Bok, D.; Sun, H. A membrane receptor for retinol binding protein mediates cellular uptake of vitamin A. Science 2007, 315, 820–825. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Kawaguchi, R. The Membrane Receptor for Plasma Retinol-Binding Protein, A New Type of Cell-Surface Receptor. Int. Rev. Cell Mol. Biol. 2011, 288, 1–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isken, A.; Golczak, M.; Oberhauser, V.; Hunzelmann, S.; Driever, W.; Imanishi, Y.; Palczewski, K.; von Lintig, J. RBP4 Disrupts Vitamin A Uptake Homeostasis in a STRA6-Deficient Animal Model for Matthew-Wood Syndrome. Cell Metab. 2008, 7, 258–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collins, M.D.; Mao, G.E. Teratology of Retinoids. Annu. Rev. Pharmacol. Toxicol. 1999, 39, 399–430. [Google Scholar] [CrossRef] [PubMed]
- Matt, N.; Dupé, V.; Garnier, J.-M.; Dennefeld, C.; Chambon, P.; Mark, M.; Ghyselinck, N.B. Retinoic acid-dependent eye morphogenesis is orchestrated by neural crest cells. Development 2005, 132, 4789–4800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cvekl, A.; Tamm, E.R. Anterior eye development and ocular mesenchyme: New insights from mouse models and human diseases. BioEssays 2004, 26, 374–386. [Google Scholar] [CrossRef] [PubMed]
- Semerci, C.N.; Kalay, E.; Yıldırım, C.; Dinçer, T.; Ölmez, A.; Toraman, B.; Koçyiğit, A.; Bulgu, Y.; Okur, V.; Şatıroğlu-Tufan, L.; et al. Novel splice-site and missense mutations in the ALDH1A3 gene underlying autosomal recessive anophthalmia/microphthalmia. Br. J. Ophthalmol. 2014, 98, 832–840. [Google Scholar] [CrossRef]
- Duester, G. Keeping an eye on retinoic acid signaling during eye development. Chem. Biol. Interact. 2009, 178, 178–181. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, R.; Shintani, T.; Sakuta, H.; Kato, A.; Ohkawara, T.; Osumi, N.; Noda, M. Identification of RALDH-3, a novel retinaldehyde dehydrogenase, expressed in the ventral region of the retina. Mech. Dev. 2000, 98, 37–50. [Google Scholar] [CrossRef] [Green Version]
- Dupé, V.; Matt, N.; Garnier, J.-M.; Chambon, P.; Mark, M.; Ghyselinck, N.B. A newborn lethal defect due to inactivation of retinaldehyde dehydrogenase type 3 is prevented by maternal retinoic acid treatment. Proc. Natl. Acad. Sci. USA 2003, 100, 14036–14041. [Google Scholar] [CrossRef] [Green Version]
- Zuber, M.E. Eye Field Specification in Xenopus laevis. Curr. Top. Dev. Biol. 2010, 93, 29–60. [Google Scholar] [CrossRef] [PubMed]
- Slijkerman, R.W.N.; Song, F.; Astuti, G.D.N.; Huynen, M.A.; van Wijk, E.; Stieger, K.; Collin, R.W.J. The pros and cons of vertebrate animal models for functional and therapeutic research on inherited retinal dystrophies. Prog. Retin. Eye Res. 2015, 48, 137–159. [Google Scholar] [CrossRef] [PubMed]
- Lwigale, P.Y. Corneal Development: Different Cells from a Common Progenitor. Prog. Mol. Biol. Transl. Sci. 2015, 134, 43–59. [Google Scholar] [CrossRef] [PubMed]
- Zieske, J.D. Corneal development associated with eyelid opening. Int. J. Dev. Biol. 2004, 48, 903–911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirschi, K.K.; Li, S.; Roy, K. Induced pluripotent stem cells for regenerative medicine. Annu. Rev. Biomed. Eng. 2014, 16, 277–294. [Google Scholar] [CrossRef] [PubMed]
- Walmsley, G.; Hyun, J.; McArdle, A.; Senarath-Yapa, K.; Hu, M.; Chung, M.; Wong, V.; Longaker, M.; Wan, D. Induced Pluripotent Stem Cells in Regenerative Medicine and Disease Modeling. Curr. Stem Cell Res. Ther. 2014, 9, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Hung, S.S.C.; Khan, S.; Lo, C.Y.; Hewitt, A.W.; Wong, R.C.B. Drug discovery using induced pluripotent stem cell models of neurodegenerative and ocular diseases. Pharmacol. Ther. 2017, 177, 32–43. [Google Scholar] [CrossRef]
- Llonch, S.; Carido, M.; Ader, M. Organoid technology for retinal repair. Dev. Biol. 2018, 433, 132–143. [Google Scholar] [CrossRef]
- Kim, S.; Lowe, A.; Dharmat, R.; Lee, S.; Owen, L.A.; Wang, J.; Shakoor, A.; Li, Y.; Morgan, D.J.; Hejazi, A.A.; et al. Generation, transcriptome profiling, and functional validation of cone-rich human retinal organoids. Proc. Natl. Acad. Sci. USA 2019, 201901572. [Google Scholar] [CrossRef]
- Welby, E.; Lakowski, J.; Di Foggia, V.; Budinger, D.; Gonzalez-Cordero, A.; Lun, A.T.L.; Epstein, M.; Patel, A.; Cuevas, E.; Kruczek, K.; et al. Isolation and Comparative Transcriptome Analysis of Human Fetal and iPSC-Derived Cone Photoreceptor Cells. Stem Cell Rep. 2017, 9, 1898–1915. [Google Scholar] [CrossRef] [Green Version]
- Kaewkhaw, R.; Kaya, K.D.; Brooks, M.; Homma, K.; Zou, J.; Chaitankar, V.; Rao, M.; Swaroop, A. Transcriptome Dynamics of Developing Photoreceptors in Three-Dimensional Retina Cultures Recapitulates Temporal Sequence of Human Cone and Rod Differentiation Revealing Cell Surface Markers and Gene Networks. Stem Cells 2015, 33, 3504–3518. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, M.A.; Knoblich, J.A. Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 2014, 345, 1247125. [Google Scholar] [CrossRef] [PubMed]
- Krebs, M.P.; Collin, G.B.; Hicks, W.L.; Yu, M.; Charette, J.R.; Shi, L.Y.; Wang, J.; Naggert, J.K.; Peachey, N.S.; Nishina, P.M. Mouse models of human ocular disease for translational research. PLoS ONE 2017, 12, e0183837. [Google Scholar] [CrossRef] [PubMed]
- Chaitankar, V.; Karakülah, G.; Ratnapriya, R.; Giuste, F.O.; Brooks, M.J.; Swaroop, A. Next generation sequencing technology and genomewide data analysis: Perspectives for retinal research. Prog. Retin. Eye Res. 2016, 55, 1–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, S.; Chatterjee, S.; Mukherjee, A.; Mutsuddi, M. Whole exome sequencing: Uncovering causal genetic variants for ocular diseases. Exp. Eye Res. 2017, 164, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Langouet-Astrie, C.J.; Meinsen, A.L.; Grunwald, E.R.; Turner, S.D.; Enke, R.A. RNA sequencing analysis of the developing chicken retina. Sci. Data 2016, 3, 160117. [Google Scholar] [CrossRef] [PubMed]
- Farkas, M.H.; Au, E.D.; Sousa, M.E.; Pierce, E.A. RNA-Seq: Improving Our Understanding of Retinal Biology and Disease. Cold Spring Harb. Perspect. Med. 2015, 5, a017152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez, S.R.; Gay, M.S.; Zhang, L. Epigenetic mechanisms in heart development and disease. Drug Discov. Today 2015, 20, 799–811. [Google Scholar] [CrossRef] [Green Version]
- Corso-Díaz, X.; Jaeger, C.; Chaitankar, V.; Swaroop, A. Epigenetic control of gene regulation during development and disease: A view from the retina. Prog. Retin. Eye Res. 2018, 65, 1–27. [Google Scholar] [CrossRef]
- Hiler, D.; Chen, X.; Hazen, J.; Kupriyanov, S.; Carroll, P.A.; Qu, C.; Xu, B.; Johnson, D.; Griffiths, L.; Frase, S.; et al. Quantification of Retinogenesis in 3D Cultures Reveals Epigenetic Memory and Higher Efficiency in iPSCs Derived from Rod Photoreceptors. Cell Stem Cell 2015, 17, 101–115. [Google Scholar] [CrossRef] [Green Version]
- Kubota, T.; Miyake, K.; Hariya, N.; Mochizuki, K. Epigenetics as a basis for diagnosis of neurodevelopmental disorders: Challenges and opportunities. Expert Rev. Mol. Diagn. 2014, 14, 685–697. [Google Scholar] [CrossRef] [PubMed]
- Cvekl, A.; Mitton, K.P. Epigenetic regulatory mechanisms in vertebrate eye development and disease. Heredity (Edinb). 2010, 105, 135–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berdasco, M.; Gómez, A.; Rubio, M.J.; Català-Mora, J.; Zanón-Moreno, V.; Lopez, M.; Hernández, C.; Yoshida, S.; Nakama, T.; Ishikawa, K.; et al. DNA Methylomes Reveal Biological Networks Involved in Human Eye Development, Functions and Associated Disorders. Sci. Rep. 2017, 7, 11762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walsh, C.T.; Garneau-Tsodikova, S.; Gatto, G.J. Protein Posttranslational Modifications: The Chemistry of Proteome Diversifications. Angew. Chem, Int. Ed. 2005, 44, 7342–7372. [Google Scholar] [CrossRef] [PubMed]
- Eastlake, K.; Heywood, W.E.; Tracey-White, D.; Aquino, E.; Bliss, E.; Vasta, G.R.; Mills, K.; Khaw, P.T.; Moosajee, M.; Limb, G.A. Comparison of proteomic profiles in the zebrafish retina during experimental degeneration and regeneration. Sci. Rep. 2017, 7, 44601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh-Roy, A.; Goncharov, A.; Jin, Y.; Chisholm, A.D. Kinesin-13 and Tubulin Posttranslational Modifications Regulate Microtubule Growth in Axon Regeneration. Dev. Cell 2012, 23, 716–728. [Google Scholar] [CrossRef] [Green Version]
- Tan, S.Z.; Begley, P.; Mullard, G.; Hollywood, K.A.; Bishop, P.N. Introduction to metabolomics and its applications in ophthalmology. Eye 2016, 30, 773–783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prosser, G.A.; Larrouy-Maumus, G.; de Carvalho, L.P.S. Metabolomic strategies for the identification of new enzyme functions and metabolic pathways. EMBO Rep. 2014, 15, 657–669. [Google Scholar] [CrossRef]
- Klupczyńska, A.; Dereziński, P.; Kokot, Z.J. Metabolomics in Medical Sciences--Trends, Challenges and Perspectives. Acta Pol. Pharm. 2015, 72, 629–641. [Google Scholar]
- Wang, X.; Gregory-Evans, K.; Wasan, K.M.; Sivak, O.; Shan, X.; Gregory-Evans, C.Y. Efficacy of Postnatal In Vivo Nonsense Suppression Therapy in a Pax6 Mouse Model of Aniridia. Mol. Ther. Nucleic Acids 2017, 7, 417–428. [Google Scholar] [CrossRef] [Green Version]


| Gene Name | OMIM# | Disease Name | Disease OMIM# | Anophthalmia/Microphthalmia (A/M) | Isolated/Complex (I/C) (Associated Ocular Features) | Non-Syndromic/Syndromic (N/S) | Reference(s) | |
|---|---|---|---|---|---|---|---|---|
| 1 | SOX2 | 184429 | Microphthalmia, syndromic 3 (MCOPS3) | 206900 | A, M | I, C (coloboma, microcornea, iris defect, retinal tuft, optic nerve hypoplasia, reduced palpebral fissure, congenital cataract, glaucoma, colobomatous cyst, synechiae, anterior segment dysgenesis, retinal/chorioretinal dystrophy, myopia) | N, S | [7,10,19,89,90] |
| 2 | OTX2 | 600037 | Microphthalmia, syndromic 5 (MCOPS5) | 610125 | A, M | I, C (coloboma, microcornea, retinal defect, optic nerve hypoplasia/aplasia, small/absent optic chiasm, LCA, early onset retinal dystrophy, hyperopia, amblyopia, cataract, focal retinal dysplasia, corectopia, synechiae, sclerocornea, persistent pupillary membrane, nystagmus, posterior vitreous opacity) | N, S | [7,18,19,21,80,90,91,92,93] |
| 3 | RAX | 601881 | Microphthalmia, isolated 3 (MCOP3) | 611038 | A, M | I, C (coloboma, sclerocornea, persistent fetal vasculature, retinal detachment, optic nerve atrophy/hypoplasia) | N, S | [7,90,94] |
| 4 | VSX2 | 142993 | Microphthalmia, isolated 2 (MCOP2) | 610093 | A, M | I, C (coloboma, congenital cataract/cloudy cornea, iris defect, microcornea, no pupillary aperture, retinal detachment, dislocated lens, small/underdeveloped optic nerve/chiasm, retinal dysfunction) | N, S | [7,66,95] |
| Microphthalmia, isolated with coloboma 3 (MCOPCB3) | 610092 | |||||||
| 5 | PAX6 | 607108 | Aniridia 1 (AN1) | 106210 | A, M | I, C (coloboma, aniridia/iris hypoplasia, anterior segment dysgenesis, agenesis of optic nerve/chiasm, primary aphakia, sclerocornea, congenital glaucoma) | N, S | [7,92] |
| 6 | STRA6 | 610745 | Microphthalmia, syndromic 9 (MCOPS9) | 601186 | A, M | I, C (coloboma, cyst, retinal detachment, abnormal cornea/iris) | N, S | [19,20,21,96,97,98] |
| 7 | RARβ | 180220 | Microphthalmia, syndromic 12 (MCOPS12) | 615524 | A, M | I, C (coloboma, sclerocornea, anterior segment dysgenesis) | S | [21,87,88] |
| 8 | ALDH1A3 | 600463 | Microphthalmia, isolated 8 (MCOP8) | 615113 | A, M | I, C (coloboma, microcornea corectopia, cyst, hypoplastic/small optic nerve/tract/chiasm, small/short palpebral fissure, conjunctival discoloration, symblepharon, nystagmus, iris attachment to the cornea) | N, S | [99,100,101] |
| 9 | FOXE3 | 601094 | Anterior segment dysgenesis 2 (ASD2) | 610256 | M | C (coloboma, anterior segment dysgenesis, sclerocornea, aphakia, aniridia) | N, S | [7] |
| 10 | BMP4 | 112262 | Microphthalmia, syndromic 6 (MCOPS6) | 607932 | A, M | I, C (coloboma, microcornea, retinal dystrophy, myopia, sclerocornea, anterior segment dysgenesis, corectopia, blepharophimosis, optic nerve hypoplasia, tilted/anomalous optic disc, cyst, nystagmus, cataract, glaucoma, aphakia, embryotoxon, persistent hypoplastic primary vitreous) | N, S | [102,103] |
| 11 | BMP7 | 112267 | - | - | A, M | I, C (coloboma) | S | [104] |
| 12 | GDF3 | 606522 | Microphthalmia, isolated 7 (MCOP7) | 613704 | M | I, C (coloboma, optic nerve hypoplasia, foveal hypoplasia, nystagmus) | N, S | [105] |
| Microphthalmia, isolated with coloboma 6 (MCOPCB6) | 613703 | |||||||
| 13 | GDF6 | 601147 | Microphthalmia, isolated 4 (MCOP4) | 613094 | A, M | I, C (coloboma, optic nerve hypoplasia, foveal hypoplasia, nystagmus) | N, S | [7,21,90] |
| Microphthalmia, isolated with coloboma 6, digenic (MCOPCB6) | 613703 | |||||||
| 14 | ABCB6 | 605452 | Microphthalmia, isolated with coloboma 7 (MCOPCB7) | 614497 | M | C (coloboma) | N | [106] |
| 15 | ATOH7 | 609875 | Persistent hyperplastic primary vitreous, autosomal recessive (PHPVAR) | 221900 | M | I, C (microcornea, congenital cataract/corneal opacity, optic nerve aplasia/hypoplasia, retinal detachment/nonattachment, persistent fetal vasculature, nystagmus, vitreous degeneration, glaucoma, shallow anterior chamber, anterior displacement of the iris, peripheral anterior synechiae, calcifications present on the hyaloid membranes/retina/vitreous, vitreoretinal dysplasia) | N | [107] |
| 16 | C12orf57 | 615140 | Temtamy syndrome (TEMTYS) | 218340 | M | C (coloboma) | S | [108] |
| 17 | TENM3 | 610083 | Microphthalmia, isolated with coloboma 9 (MCOPCB9) | 615145 | M | C (coloboma, microcornea, nystagmus, esotropia, myopia, retinal detachment) | N, S | [109,110] |
| 18 | VAX1 | 604294 | Microphthalmia, syndromic 11 (MCOPS11) | 614402 | A, M | C (optic nerve hypoplasia, small optic nerve, cyst) | S | [111] |
| 19 | SMOC1 | 608488 | Microphthalmia with limb anomalies (MLA) | 206920 | A | C (optic nerve/tract aplasia) | S | [112] |
| 20 | FNBP4 | 615265 | Microphthalmia with limb anomalies (MLA) | 206920 | A | I | S | [113] |
| 21 | SHH | 600725 | Microphthalmia, isolated with coloboma 5 (MCOPCB5) | 611638 | A, M | I, C (coloboma, funnel retinal detachment with sub-retinal opacity, microcornea, small optic nerve, retinal dystrophy, tilted optic disc, myopia, nystagmus, glaucoma, posterior embryotoxon) | N, S | [102,114,115] |
| 22 | NAA10 | 300013 | Microphthalmia, syndromic 1 (MCOPS1) | 309800 | A | I | S | [116] |
| 23 | BCOR | 300485 | Microphthalmia, syndromic 2 (MCOPS2) | 300166 | A, M | I, C (congenital cataract, microcornea, posterior embryotoxon, secondary aphakia, secondary glaucoma, retinal detachment, persistent fetal vasculature, iris heterochromia, nystagmus, myopia, iris rubeosis, flat anterior chambers) | S | [117] |
| 24 | HCCS | 300056 | Linear skin defects with multiple congenital anomalies 1 (LSDMCA1) | 309801 | A, M | I, C (corneal opacity/cloudy and vascular cornea, cyst, sclerocornea, glaucoma,) | S | [118] |
| 25 | MAB21L2 | 604357 | Microphthalmia, syndromic 14 (MCSKS) | 615877 | A, M | I, C (coloboma, microcornea, exotropia, sclerocornea, strabismus) | S | [119] |
| 26 | RBP4 | 180250 | Microphthalmia, isolated with coloboma 10 (MCOPCB10) | 616428 | A, M | C (coloboma, small optic nerve/chiasm, cyst, underdeveloped extraocular muscles) | N, S | [81,120] |
| 27 | GLI2 | 165230 | Holoprosencephaly 9 (HPE9) | 610829 | A, M | I, C (coloboma, optic nerve agenesis) | S | [121,122,123] |
| 28 | PORCN | 300651 | Focal dermal hypoplasia (FDH) | 305600 | A, M | I, C (coloboma, aniridia, strabismus, ectopia lentis) | S | [124,125] |
| 29 | FRAS1 | 607830 | Fraser syndrome 1 (FRASRS1) | 219000 | A, M | C (fused/small palpebral fissure, cryptophthalmos) | S | [126] |
| 30 | FREM1 | 608944 | Manitoba oculotrichoanal syndrome (MOTA) | 248450 | A | I, C (coloboma, obstruction of the nasolacrimal duct) | S | [127] |
| 31 | SMCHD1 | 614982 | Bosma arhinia microphthalmia syndrome (BAMS) | 603457 | M | I, C (coloboma, hypertelorism, occluded or absent nasolacrimal duct, cataract) | S | [128] |
| 32 | SIX6 | 606326 | Microphthalmia, syndromic 6 (MCOPS6) | 607932 | A, M | C (coloboma, cataract, nystagmus, secondary glaucoma, optic nerve dysplasia/absence of optic nerve/chiasm/tract, retinal dystrophy, cyst) | S | [129] |
| 33 | TFAP2A | 107580 | Branchiooculofacial syndrome (BOFS) | 113620 | A, M | C (coloboma, cataract/corneal clouding, reduced corneal diameter, primary aphakia, sclerocornea, retinal detachment, lacrimal duct obstruction, cyst, subluxed cataractous lens, shallow anterior chamber, persistent pupillary membrane, iris hypoplasia, dysplastic optic disc) | S | [130] |
| 34 | TCTN2 | 613846 | Meckel syndrome, type 8 (MKS8) | 613885 | A, M | I | S | [131] |
| 35 | CSPP1 | 611654 | Joubert syndrome 21 (JBTS21) | 615636 | A | I | S | [132] |
| 36 | COL4A1 | 120130 | Brain small vessel disease with or without ocular anomalies (BSVD1) | 175780 | M | C (microcornea, Peter’s anomaly, retinal detachment, congenital cataract, glaucoma, anterior segment dysgenesis, hypermetropia, astigmatism) | S | [133,134] |
| 37 | PTCH1 | 601309 | Holoprosencephaly 7 (HPE7) | 610828 | M | C (coloboma, cataract, sclerocornea, anterior segment dysgenesis) | N, S | [135] |
| 38 | TBC1D32 | 615867 | Orofaciodigital syndrome IX (OFD9) | 258865 | A, M | C (coloboma) | S | [136] |
| 39 | CHD7 | 605806 | CHARGE syndrome | 214800 | A, M | I, C (coloboma, microcornea, cataract, persistent fetal vasculature) | N, S | [137,138,139] |
| 40 | MFRP | 606227 | Microphthalmia, isolated 5 (MCOP5) | 611040 | M | C (retinitis pigmentosa, foveoschisis, optic disc drusen, macular edema, glaucoma, hyperopia) | N | [140] |
| Nanophthalmos 2 (NN02) | 609549 | |||||||
| 41 | PRSS56 | 613858 | Microphthalmia, isolated 6 (MCOP6) | 613517 | M | C (hyperopia, elevated papillomacular retinal fold, shallow anterior chamber, thick lens, thickened scleral wall) | N | [141,142] |
| 42 | TMEM98 | 615949 | Nanophthalmos 4 (NN04) | 615972 | M | C (hyperopia, angle closure glaucoma, narrow iridocorneal angle, shallow anterior chamber depth, optic disc drusen) | N | [143,144] |
| 43 | HMGB3 | 300193 | Microphthalmia, syndromic 13 (MCOPS13) | 300915 | A, M | C (coloboma, congenital cataract) | S | [145] |
| 44 | PXDN | 605158 | Cornea opacification and other ocular anomalies (ASGD7) | 269400 | M | C (sclerocornea, anterior segment dysgenesis, iridocorneal dysgenesis, glaucoma, cataract) | N, S | [146] |
| 45 | TMX3 | 616102 | Microphthalmia with coloboma 1 (MCOPCB1) | 300345 | A, M | I, C (coloboma, cyst) | N, S | [147] |
| 46 | YAP1 | 606608 | Coloboma, ocular, with or without hearing impairment, cleft lip/palate, and/or mental retardation (COB1) | 120433 | A, M | I, C (coloboma, extraocular muscle defect, cataract, ectopic pupil) | N, S | [148] |
| 47 | IPO13 | 610411 | - | - | M | C (coloboma, cataract, narrowed palpebral fissure, nystagmus, microcornea) | N | [149] |
| 48 | PITX3 | 602669 | Cataract 11, multiple types (CTRCT11) | 610623 | M | C (cataract/corneal opacity) | S | [150] |
| 49 | NDP | 300658 | Norrie disease (ND) | 310600 | M | C (sclerocornea) | N | [92] |
| 50 | MITF | 156845 | COMMAD syndrome | 617306 | M | C (coloboma, microcornea with pannus, cataract, translucent irides, optic nerve/tract hypoplasia) | S | [151] |
| 51 | FOXC1 | 601090 | - | - | M | C (microcornea, sclerocornea, cyst, myopia, cataract, Rieger anomaly, retinal detachment) | N | [152] |
| 52 | CRPPA | 614631 | Muscular dystrophy-dystroglycanopathy (congenital with brain and eye anomalies), type A, 7 (MDDGA7) | 614643 | M | C (cataract, optic nerve hypoplasia) | S | [153] |
| 53 | FANCL | 608111 | Fanconi anemia, complementation group L (FANCL) | 614083 | M | C (short upslant palpebral fissure, indiscernible pupil) | S | [154] |
| 54 | SMO | 601500 | Curry-Jones syndrome (CRJS) | 601707 | M | C (coloboma, unusually shaped pupil) | S | [155] |
| 55 | DOCK6 | 614194 | Adams-Oliver syndrome 2 (AOS2) | 614219 | M | I, C (retinal detachment) | S | [156] |
| 56 | CRYAA | 123580 | Cataract 9, multiple types (CTRCT9) | 604219 | M | C (congenital cataract) | N | [157] |
| 57 | FOXL2 | 605597 | Blepharophimosis, ptosis and epicanthus inversus (BPES) | 110100 | M | C (blepharophimosis, ptosis, epicanthus inversus, telecanthus, strabismus) | N, S | [158] |
| 58 | CRYBA4 | 123631 | Cataract 23, multiple types (CTRCT23) | 610425 | M | C (cataract, enophthalmia) | N | [159] |
| 59 | ERCC6 | 609413 | Cerebrooculofacioskeletal syndrome 1 (COFS1) | 214150 | M | C (congenital cataract, short palpebral fissure, blepharokeratoconjunctivitis) | S | [160,161] |
| 60 | ERCC5 | 133530 | Cerebrooculofacioskeletal syndrome 3 (COFS3) | 616570 | M | C (cataract) | S | [162] |
| 61 | ERCC1 | 126380 | Cerebrooculofacioskeletal syndrome 4 (COFS4) | 610758 | M | C (blepharophimosis) | S | [163] |
| 62 | SRD5A3 | 611715 | Congenital disorder of glycosylation, type 1q (CDG1q) | 612379 | M | C (coloboma, nystagmus, cataract, optic atrophy) | S | [164] |
| 63 | SALL4 | 607343 | Duane-radial ray syndrome (DRRS) | 607323 | M | C (coloboma, optic nerve hypoplasia) | S | [165] |
| 64 | FREM2 | 608945 | Fraser syndrome 2 (FRASRS2) | 617666 | M | C (coloboma, cyst) | S | [166] |
| 65 | RPGRIP1L | 610937 | Meckel syndrome 5 (MKS5) | 611561 | M | I | S | [167] |
| 66 | SLC25A24 | 608744 | Fontaine progeroid syndrome (FPS) | 612289 | M | I | S | [168] |
| 67 | FAM111A | 615292 | Gracile bone dysplasia (GCLEB) | 602361 | M | I | S | [169] |
| 68 | SMG9 | 613176 | Heart and brain malformation syndrome (HBMS) | 616920 | M | I | S | [170] |
| 69 | SIX3 | 603714 | Holoprosencephaly 2 (HPE2) | 157170 | M | C (coloboma, myopia, astigmatism, dysplastic optic nerve, nystagmus, exotropia, cataract, hypertropia) | S | [171] |
| 70 | PDE6D | 602676 | Joubert syndrome 22 (JBTS22) | 615665 | M | I, C (coloboma) | S | [172] |
| 71 | KMT2D | 602113 | Kabuki syndrome 1 (KABUK1) | 147920 | M | C (cyst) | S | [173] |
| 72 | PAX2 | 167409 | Papillorenal syndrome (PAPRS) | 120330 | M | C (coloboma, optic nerve dysplasia, retinal degeneration) | S | [174,175] |
| 73 | TMEM216 | 613277 | Meckel syndrome, type 2 (MKS2) | 603194 | M | I | S | [176] |
| 74 | CEP290 | 610142 | Meckel syndrome, type 4 (MKS4) | 611134 | M | I | S | [177] |
| 75 | KIF11 | 148760 | Microcephaly with or without chorioretinopathy, lymphedema, or mental retardation (MCLMR) | 152950 | M | I, C (coloboma, cataract, chorioretinopathy, hypermetropia, persistent hyaloid artery, peripheral fibrovascular proliferation, retinal detachment) | S | [178,179] |
| 76 | SNX3 | 605930 | - | - | M | I | S | [180] |
| 77 | ZEB2 | 605802 | Mowat-Wilson syndrome (MOWS) | 235730 | M | C (cataract, retinal aplasia, corectopia, optic nerve hypoplasia/pallor, retinal atrophy) | S | [181] |
| 78 | POMT1 | 607423 | Muscular dystrophy-dystroglycanopathy (congenital with brain and eye anomalies), type A, 1 (MDDGA1) | 236670 | M | C (anterior chamber dysgenesis, exophthalmia, buphthalmos, megalocornea, glaucoma, retinal dysplasia, congenital cataract/corneal clouding, retinal detachment) | S | [182,183] |
| 79 | POMT2 | 607439 | Muscular dystrophy-dystroglycanopathy (congenital with brain and eye anomalies), type A, 2 (MDDGA2) | 613150 | M | C (Peter’s anomaly, cataract, buphthalmos) | S | [184] |
| 80 | POMGNT1 | 614828 | - | - | M | C (corneal clouding/cataract) | S | [185] |
| 81 | FKTN | 607440 | Muscular dystrophy-dystroglycanopathy (congenital with brain and eye anomalies), type A, 4 (MDDGA4) | 253800 | M | C (retinal detachment) | S | [186] |
| 82 | FKRP | 606596 | Muscular dystrophy-dystroglycanopathy (congenital with brain and eye anomalies), type A, 5 (MDDGA5) | 613153 | M | C (cataract, asymmetric pupils, persistent hyperplastic primary vitreous, anterior chamber abnormality) | S | [187,188] |
| 83 | DAG1 | 128239 | Muscular dystrophy-dystroglycanopathy (congenital with brain and eye anomalies), type A, 9 (MDDGA9) | 616538 | M | C (buphthalmos, corneal opacity, glaucoma, retinal detachment) | S | [189,190] |
| 84 | B3GALNT2 | 610194 | Muscular dystrophy-dystroglycanopathy (congenital with brain and eye anomalies, type A, 11 (MDDGA11) | 615181 | M | C (cataract, optic nerve hypoplasia, myopia) | S | [191] |
| 85 | RAB3GAP1 | 602536 | Warburg micro syndrome 1 (WARBM1) | 600118 | M | C (microcornea, cataract) | S | [192] |
| 86 | RAB3GAP2 | 609275 | Warburg micro syndrome 2 (WARBM2) | 614225 | M | C (congenital cataract, small pupil, aphakia, hypermetropia, secondary glaucoma) | S | [193,194] |
| Martsolf syndrome | 212720 | |||||||
| 87 | NHS | 300457 | Nance-Horan syndrome (NHS) | 300672 | M | C (microcornea, congenital cataract) | S | [195] |
| Cataract 40, X-linked (CTRCT40) | 302200 | |||||||
| 88 | HMX1 | 142992 | Oculoauricular syndrome (OCACS) | 612109 | M | C (microcornea, coloboma, nystagmus, cataract, microphakia, synechiae, anterior segment dysgenesis, small dysplastic optic disc, strabismus, sclerocornea, posterior embryotoxon, stromal iris cyst, retinal dystrophy, dysplastic macropapillae, macular hypoplasia, irido-corneal adherences, enophthalmus, esotropia, calcified phthisis bulbi) | S | [196,197] |
| 89 | GJA1 | 121014 | Oculodentodigital dysplasia, autosomal recessive | 257850 | M | I, C (cataract, uveitis, glaucoma, persistent pupillary membrane) | S | [198,199,200] |
| Oculodentodigital dysplasia (ODDD) | 164200 | |||||||
| 90 | LRP5 | 603506 | Osteoporosis-pseudoglioma syndrome (OPPG) | 259770 | M | C (retinal detachment, persistent hyperplasia of the primary vitreous) | S | [201] |
| 91 | PQBP1 | 300463 | Renpenning syndrome (RENS1) | 309500 | M | C (coloboma) | S | [202] |
| 92 | TUBB | 191130 | Symmetric circumferential skin creases, congenital, 1 (CSCSC1) | 156610 | M | C (short palpebral fissure, retinal dysplasia, microcornea) | S | [203,204] |
| Cortical dysplasia, complex, with other brain malformations 6 (CDCBM6) | 615771 | M | ||||||
| 93 | MAPRE2 | 605789 | Symmetric circumferential skin creases, congenital, 2 (CSCSC2) | 616734 | M | C (short/slanting palpebral fissure, strabismus, ptosis) | S | [203] |
| 94 | SALL1 | 602218 | Townes-Brocks syndrome 1 (TBS1) | 107480 | A, M | C (abnormal lens, aplastic optic nerve, small optic chiasm) | S | [205] |
| 95 | HDAC6 | 300272 | Chondrodysplasia with platyspondyly, distinctive brachydactyly, hydrocephaly, and microphthalmia | 300863 | M | I | S | [206,207] |
| 96 | ALX1 | 601527 | Frontonasal dysplasia 3 (FND3) | 613456 | M | C (coloboma) | S | [208] |
| 97 | RERE | 605226 | Neurodevelopmental disorder with or without anomalies of the brain, eye, or heart (NEDBEH) | 616975 | M | C (coloboma, optic nerve hypoplasia, anisometropia) | S | [209] |
| 98 | RAB18 | 602207 | Warburg micro syndrome 3 | 614222 | M | C (microcornea, congenital cataract, small atonic pupil, progressive optic atrophy) | S | [210] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harding, P.; Moosajee, M. The Molecular Basis of Human Anophthalmia and Microphthalmia. J. Dev. Biol. 2019, 7, 16. https://0-doi-org.brum.beds.ac.uk/10.3390/jdb7030016
Harding P, Moosajee M. The Molecular Basis of Human Anophthalmia and Microphthalmia. Journal of Developmental Biology. 2019; 7(3):16. https://0-doi-org.brum.beds.ac.uk/10.3390/jdb7030016
Chicago/Turabian StyleHarding, Philippa, and Mariya Moosajee. 2019. "The Molecular Basis of Human Anophthalmia and Microphthalmia" Journal of Developmental Biology 7, no. 3: 16. https://0-doi-org.brum.beds.ac.uk/10.3390/jdb7030016






