Aggregate Removal Nanofiltration of Human Serum Albumin Solution Using Nanocellulose-Based Filter Paper
Abstract
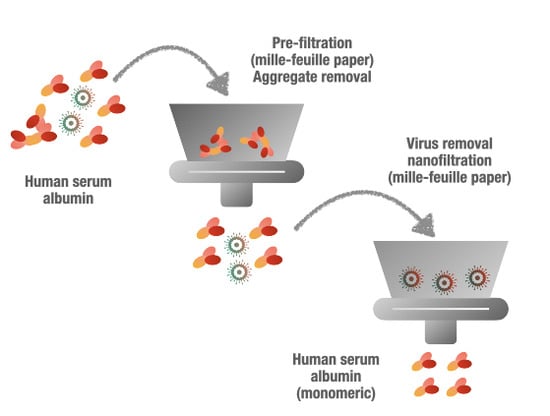
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
Filter Paper Preparation
2.3. Filtration
2.3.1. Filtration Setup
2.3.2. Prefiltration
2.3.3. Virus Removal Filtration
2.3.4. Vmax Analysis
2.3.5. Scanning Electron Microscopy
2.3.6. Nitrogen Gas Sorption
2.3.7. Dynamic Light Scattering
2.3.8. Size Exclusion High Performance Liquid Chromatography (SE-HPLC)
2.3.9. Protein Recovery
2.3.10. Plaque Forming Units (PFU) and log10 Reduction Value (LRV)
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| BSA | bovine serum albumin |
| CHO | Chinese hamster ovary cells |
| CPV | canine parvovirus |
| DLS | dynamic light scattering |
| DMEM | Dulbecco’s modified Eagle’s medium |
| HSA | human serum albumin |
| ITS | insulin-transferrin-selenium |
| IVIG | intravenous immunoglobulin |
| LBM | Luria-Bertani medium |
| LRV | log10 reduction value |
| MVM | minute virus of mice |
| PFU | plaque forming units |
| PBS | phosphate buffer saline |
| SE-HPLC | size-exclusion high performance liquid chromatography |
| SEM | scanning electron microscopy |
| xMuLV | xenotropic murine leukemia virus |
References
- Doweiko, J.P.; Nompleggi, D.J. Reviews: Role of Albumin in Human Physiology and Pathophysiology. J. Parenter. Enter. Nutr. 1991, 15, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Elsadek, B.; Kratz, F. Impact of albumin on drug delivery—New applications on the horizon. J. Control. Release 2012, 157, 4–28. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ueno, H.; Masuko, T.; Hashimoto, Y. Binding of Serum Albumin on Tumor Cells and Characterization of the Albumin Binding Protein. J. Biochem. 1994, 115, 898–903. [Google Scholar] [CrossRef] [PubMed]
- De Castro, M.; Orive, G.; Gascón, A.; Hernandez, R.; Pedraz, J. Evaluation of human serum albumin as a substitute of foetal bovine serum for cell culture. Int. J. Pharm. 2006, 310, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Hreinsson, J.; Zhang, P.; Swahn, M.L.; Hultenby, K.; Hovatta, O. Cryopreservation of follicles in human ovarian cortical tissue. Comparison of serum and human serum albumin in the cryoprotectant solutions. Hum. Reprod. 2003, 18, 2420–2428. [Google Scholar] [CrossRef]
- Goyen, M. Gadofosveset-enhanced magnetic resonance angiography. Vasc. Health Risk Manag. 2008, 4, 1. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, K. Summary of recombinant human serum albumin development. Biologicals 2006, 34, 55–59. [Google Scholar] [CrossRef]
- He, Y.; Ning, T.; Xie, T.; Qiu, Q.; Zhang, L.; Sun, Y.; Jiang, D.; Fu, K.; Yin, F.; Zhang, W. Large-scale production of functional human serum albumin from transgenic rice seeds. Proc. Natl. Acad. Sci. USA 2011, 108, 19078–19083. [Google Scholar] [CrossRef] [Green Version]
- Anraku, M.; Tsurusaki, Y.; Watanabe, H.; Maruyama, T.; Kragh-Hansen, U.; Otagiri, M. Stabilizing mechanisms in commercial albumin preparations: Octanoate and N-acetyl-l-tryptophanate protect human serum albumin against heat and oxidative stress. Biochim. et Biophys. Acta (BBA)-Proteins Proteom. 2004, 1702, 9–17. [Google Scholar] [CrossRef]
- Anraku, M.; Kouno, Y.; Kai, T.; Tsurusaki, Y.; Yamasaki, K.; Otagiri, M. The role of N-acetyl-methioninate as a new stabilizer for albumin products. Int. J. Pharm. 2007, 329, 19–24. [Google Scholar] [CrossRef]
- Kouno, Y.; Anraku, M.; Yamasaki, K.; Okayama, Y.; Iohara, D.; Nakamura, H.; Maruyama, T.; Hirayama, F.; Kragh-Hansen, U.; Otagiri, M. N-acetyl-L-methionine is a superior protectant of human serum albumin against post-translational oxidation as compared to N-acetyl-L-tryptophan. Biochem. Biophys. Rep. 2016, 6, 266–274. [Google Scholar] [CrossRef] [Green Version]
- Erstad, B.L. Viral infectivity of albumin and plasma protein fraction. Pharmacother. J. Hum. Pharmacol. Drug Ther. 1996, 16, 996–1001. [Google Scholar]
- Blümel, J.; Nhan, D.M.T.-X.; Teitz, S.; Miyabayashi, T.; Boller, K.; Schnierle, B.S.; Baylis, S.A. Inactivation and removal of Zika virus during manufacture of plasma-derived medicinal products. Transfusion 2016, 57, 790–796. [Google Scholar] [CrossRef] [Green Version]
- Farcet, M.R.; Kreil, T.R. Zika virus is not thermostable: Very effective virus inactivation during heat treatment (pasteurization) of human serum albumin. Transfusion 2016, 57, 797–801. [Google Scholar] [CrossRef] [PubMed]
- Kühnel, D.; Müller, S.; Pichotta, A.; Radomski, K.U.; Volk, A.; Schmidt, T. Inactivation of Zika virus by solvent/detergent treatment of human plasma and other plasma-derived products and pasteurization of human serum albumin. Transfusion 2016, 57, 802–810. [Google Scholar] [CrossRef] [PubMed]
- Leydold, S.M.; Farcet, M.R.; Kindermann, J.; Modrof, J.; Pölsler, G.; Berting, A.; Howard, M.K.; Barrett, P.N.; Kreil, T.R. Chikungunya virus and the safety of plasma products. Transfusion 2012, 52, 2122–2130. [Google Scholar] [CrossRef] [PubMed]
- Yue, C.; Teitz, S.; Miyabashi, T.; Boller, K.; Lewis-Ximenez, L.L.; Baylis, S.A.; Blümel, J. Inactivation and Removal of Chikungunya Virus and Mayaro Virus from Plasma-derived Medicinal Products. Viruses 2019, 11, 234. [Google Scholar] [CrossRef] [Green Version]
- Blümel, J.; Rinckel, L.A.; Lee, D.C.; Roth, N.J.; Baylis, S.A. Inactivation and neutralization of parvovirus B19 Genotype 3. Transfusion 2012, 52, 1490–1497. [Google Scholar] [CrossRef]
- Blümel, J.; Schmidt, I.; Willkommen, H.; Löwer, J. Inactivation of parvovirus B19 during pasteurization of human serum albumin. Transfusion 2002, 42, 1011–1018. [Google Scholar] [CrossRef]
- Farcet, M.R.; Kindermann, J.; Modrof, J.; Kreil, T.R. Inactivation of hepatitis A variants during heat treatment (pasteurization) of human serum albumin. Transfusion 2011, 52, 181–187. [Google Scholar] [CrossRef]
- Gröner, A.; Broumis, C.; Fang, R.; Nowak, T.; Popp, B.; Schäfer, W.; Roth, N.J. Effective inactivation of a wide range of viruses by pasteurization. Transfusion 2017, 58, 41–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klammt, S.; Brinkmann, B.; Mitzner, S.; Munzert, E.; Loock, J.; Stange, J.; Emmrich, J.; Liebe, S. Albumin Binding Capacity (ABiC) is reduced in commercially available Human Serum Albumin preparations with stabilizers. Zeitschrift für Gastroenterologie 2001, 39, 24–27. [Google Scholar] [CrossRef] [PubMed]
- Harm, S.; Schildböck, C.; Hartmann, J. Removal of stabilizers from human serum albumin by adsorbents and dialysis used in blood purification. PLoS ONE 2018, 13, e0191741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bar-Or, D.; Bar-Or, R.; Rael, L.; Gardner, D.K.; Slone, D.S.; Craun, M.L. Heterogeneity and oxidation status of commercial human albumin preparations in clinical use. Crit. Care Med. 2005, 33, 1638–1641. [Google Scholar] [CrossRef]
- Stange, J. Extracorporeal liver support. Organog 2011, 7, 64–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, W.-W.; MacKenzie, A.D.; Nelson, V.J.; Faed, J.M.; Turner, P.R. Octanoate in Human Albumin Preparations Is Detrimental to Mesenchymal Stromal Cell Culture. Stem Cells Int. 2015, 2015, 192576. [Google Scholar] [CrossRef]
- Lang, B.E.; Cole, K.D. Unfolding properties of recombinant human serum albumin products are due to bioprocessing steps. Biotechnol. Prog. 2014, 31, 62–69. [Google Scholar] [CrossRef]
- Dengler, T.; Stöcker, U.; Kellner, S.; Fürst, G. Chemical and Immunochemical Characterization of Polymers or Aggregates in Preparations of Human Serum Albumin. Transfus. Med. Hemother. 1989, 16, 160–164. [Google Scholar] [CrossRef]
- Jensen, L.B.; Dam, J.; Teisner, B. Identification and Removal of Polymer- and Aggregate-Forming Proteins in Human Plasma Albumin Preparations. Vox Sang. 1994, 67, 125–131. [Google Scholar] [CrossRef]
- Gay, M.; Carrascal, M.; Gorga, M.; Parés, A.; Abian, J. Characterization of peptides and proteins in commercial HSA solutions. Proteomics 2010, 10, 172–181. [Google Scholar] [CrossRef]
- Lin, J.-J.; Meyer, J.D.; Carpenter, J.F.; Manning, M.C. Stability of human serum albumin during bioprocessing: Denaturation and aggregation during processing of albumin paste. Pharm. Res. 2000, 17, 391–396. [Google Scholar] [CrossRef]
- Lin, J.-J.; Meyer, J.D.; Carpenter, J.F.; Manning, M.C. Aggregation of human serum albumin during a thermal viral inactivation step. Int. J. Boil. Macromol. 2009, 45, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, T.; Katoh, S.; Nakajima, M.; Nabetani, H. Mechanism of bovine serum albumin aggregation during ultrafiltration. Biotechnol. Bioeng. 2001, 75, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Metreveli, G.; Wågberg, L.; Emmoth, E.; Belák, S.; Strømme, M.; Mihranyan, A. A Size-Exclusion Nanocellulose Filter Paper for Virus Removal. Adv. Healthc. Mater. 2014, 3, 1546–1550. [Google Scholar] [CrossRef] [PubMed]
- Asper, M.; Hanrieder, T.; Quellmalz, A.; Mihranyan, A. Removal of xenotropic murine leukemia virus by nanocellulose based filter paper. Biologicals 2015, 43, 452–456. [Google Scholar] [CrossRef]
- Gustafsson, S.; Lordat, P.; Hanrieder, T.; Asper, M.; Schaefer, O.; Mihranyan, A. Mille-feuille paper: A novel type of filter architecture for advanced virus separation applications. Mater. Horizons 2016, 3, 320–327. [Google Scholar] [CrossRef]
- Gustafsson, S.; Mihranyan, A. Strategies for Tailoring the Pore-Size Distribution of Virus Retention Filter Papers. ACS Appl. Mater. Interfaces 2016, 8, 13759–13767. [Google Scholar] [CrossRef]
- Manukyan, L.; Li, P.; Gustafsson, S.; Mihranyan, A. Growth media filtration using nanocellulose-based virus removal filter for upstream biopharmaceutical processing. J. Membr. Sci. 2019, 572, 464–474. [Google Scholar] [CrossRef]
- Manukyan, L.; Padova, J.; Mihranyan, A. Virus removal filtration of chemically defined Chinese Hamster Ovary cells medium with nanocellulose-based size exclusion filter. Biologicals 2019, 59, 62–67. [Google Scholar] [CrossRef]
- Wu, L.; Manukyan, L.; Mantas, A.; Mihranyan, A. Nanocellulose-Based Nanoporous Filter Paper for Virus Removal Filtration of Human Intravenous Immunoglobulin. ACS Appl. Nano Mater. 2019, 2, 6352–6359. [Google Scholar] [CrossRef]
- Manukyan, L.; Mantas, A.; Razumikhin, M.; Katalevsky, A.; Golubev, E.; Mihranyan, A. Two-Step Size-Exclusion Nanofiltration of Prothrombin Complex Concentrate Using Nanocellulose-Based Filter Paper. Biomedicines 2020, 8, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Badmington, F.; Wilkins, R.; Payne, M.; Honig, E.S. Vmax testing for practical microfiltration train scale-up in biopharmaceutical processing. Pharm. Technol. 1995, 19, 64–76. [Google Scholar]
- Barrett, E.P.; Joyner, L.G.; Halenda, P.P. The determination of pore volume and area distributions in porous substances. I. Computations from nitrogen isotherms. J. Am. Chem. Soc. 1951, 73, 373–380. [Google Scholar] [CrossRef]
- Sing, K.S.; Williams, R.T. Physisorption Hysteresis Loops and the Characterization of Nanoporous Materials. Adsorpt. Sci. Technol. 2004, 22, 773–782. [Google Scholar] [CrossRef]
- Gustafsson, S.; Westermann, F.; Hanrieder, T.; Jung, L.; Ruppach, H.; Mihranyan, A. Comparative Analysis of Dry and Wet Porometry Methods for Characterization of Regular and Cross-Linked Virus Removal Filter Papers. Membranes 2018, 9, 1. [Google Scholar] [CrossRef] [Green Version]
- Gustafsson, O.; Gustafsson, S.; Manukyan, L.; Mihranyan, A. Significance of Brownian Motion for Nanoparticle and Virus Capture in Nanocellulose-Based Filter Paper. Membranes 2018, 8, 90. [Google Scholar] [CrossRef] [Green Version]






| Sample | Pressure, Bar | HSA Recovery, % | Vmax, L m−2 |
|---|---|---|---|
| Prefiltration, 11 μm | 1 | 94.3 ± 1.5 | / |
| Virus filtration, 22 μm | 1 | 90.7 ± 6.6 | ≥1000 |
| Virus filtration, 22 μm | 3 | 92.3 ± 0.6 | 110.0 ± 2.5 |
| Virus filtration, 22 μm | 5 | 91.3 ± 3.5 | 63.6 ± 5.8 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, L.; Mantas, A.; Gustafsson, S.; Manukyan, L.; Mihranyan, A. Aggregate Removal Nanofiltration of Human Serum Albumin Solution Using Nanocellulose-Based Filter Paper. Biomedicines 2020, 8, 209. https://0-doi-org.brum.beds.ac.uk/10.3390/biomedicines8070209
Wu L, Mantas A, Gustafsson S, Manukyan L, Mihranyan A. Aggregate Removal Nanofiltration of Human Serum Albumin Solution Using Nanocellulose-Based Filter Paper. Biomedicines. 2020; 8(7):209. https://0-doi-org.brum.beds.ac.uk/10.3390/biomedicines8070209
Chicago/Turabian StyleWu, Lulu, Athanasios Mantas, Simon Gustafsson, Levon Manukyan, and Albert Mihranyan. 2020. "Aggregate Removal Nanofiltration of Human Serum Albumin Solution Using Nanocellulose-Based Filter Paper" Biomedicines 8, no. 7: 209. https://0-doi-org.brum.beds.ac.uk/10.3390/biomedicines8070209