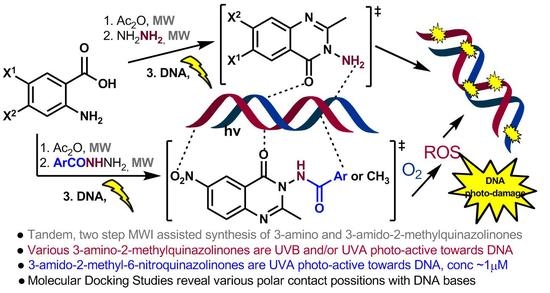
Green Process for the Synthesis of 3-Amino-2-methyl-quinazolin-4(3H)-one Synthones and Amides Thereof:DNA Photo-Disruptive and Molecular Docking Studies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Synthesis of Chemical Compounds
2.1.1. General Method for the Synthesis of Benzoxazinones (2a-2k)
2.1.2. General Method for the Synthesis of Substituted 2-methyl-3-amino–quinazolin -4(3H) ones (3a-3k)
- 3-amino-6-hydroxy-2-methylquinazolin-4(3H)-one (3a) [26]: Off white amorphous solid; mp: 294–296 °C; yield: 32%; 1H-NMR (DMSO-d6, 600 MHz): δ (ppm) 9.98 (s,1H, OH), 7.46 (d, J = 8.4 Hz, 1H, H8), 7.36 (d, J = 1.8 Hz, 1H, H5), 7.23 (dd, J = 8.4, 1.8 Hz, 1H, H7), 5.75 (s, 2H, NH2), 2.52 (s, 3H, CH3); UV-Vis (e M−1cm−1): λ (nm) = 274 (11,854), 329 (4780).
- 3-amino-2,6-dimethylquinazolin-4(3H)-one (3b) [55]: White amorphous solid; mp: 170–171 °C; yield: 31%; 1H-NMR (DMSO-d6, 600 MHz): δ (ppm) 7.89 (s, 1H, H5), 7.60 (d, J = 7.8, 1.8 Hz, 1H, H7), 7.50 (d, J = 7.8 Hz, 1H, H8), 5.78 (s, 2H, NH2), 2.56 (s, 3H, CH3), 2.44 (s, 3H, CH3); UV-Vis (e M−1cm−1): λ (nm) = 271 (5482), 312 (2268).
- 3-amino-2-methylquinazolin-4(3H)-one (3c) [56]: White amorphous solid; mp: 142–143 °C; yield: 35%; 1H-NMR (CDCl3, 300 MHz): δ (ppm) 8.08 (d, 1H, H5), 7.63–7.61 (m, 2H), 7.36–7.31 (m, 1H), 5.15 (s, 2H, NH2), 2.66 (s, 3H, CH3); UV-Vis (e M−1cm−1): λ (nm) = 273 (684).
- 3-amino-6-fluoro-2-methylquinazolin-4(3H)-one (3d) [57]: White amorphous solid; mp: 198–199 °C; yield: 51%; 1H-NMR (DMSO-d6, 600 MHz): δ (ppm) 7.76 (dd, J = 8.16, 1.6 Hz, 1H, H7), 7.68–7.66 (m, 2H, H5, H8), 5.82 (s, 2H, NH2), 2.57 (s, 3H, CH3); UV-Vis (e M−1cm−1): λ (nm) = 265 (6178), 314 (3475).
- 3-amino-6-chloro-2-methylquinazolin-4(3H)-one (3e) [58]: White amorphous solid; mp: 174–175 °C; yield: 50%; 1H-NMR (DMSO-d6, 600 MHz): δ (ppm) 8.03 (d, J = 2.3 Hz, 1H, H5), 7.80 (dd, J = 8.7, 2.4 Hz, 1H, H7), 7.63 (d, J = 8.76 Hz, 1H, H8), 5.84 (s, 2H, NH2), 2.58 (s, 3H, CH3); UV-Vis (e M−1cm−1): λ (nm) = 274 (8996), 316 (3347).
- 3-amino-7-chloro-2-methylquinazolin-4(3H)-one (3f) [59]: White amorphous solid; mp: 197–200 °C; yield: 40%; 1H-NMR (DMSO-d6, 600 MHz): δ (ppm) 8.09 (d, J = 8.5 Hz, 1H, H5), 7.39 (d, J = 1.6 Hz, 1H, H8), 7.50 (dd, J = 8.5, 1.8 Hz, 1H, H6), 5.81 (s, 2H, NH2), 2.58 (s, 3H, CH3); UV-Vis (e M−1cm−1): λ (nm) = 278 (5439).
- 3-amino-6-bromo-2-methylquinazolin-4(3H)-one (3g) [11]: Pale yellow amorphous solid; mp: 187–189 °C; yield: 70%; 1H-NMR (DMSO-d6, 600 MHz): δ (ppm) 8.15 (s, 1H, H5), 7.89 (dd, J = 8.4 Hz, 1H, H7), 7.53 (d, J = 8.6, 1H, H8), 5.83 (s, 2H, NH2), 2.57(s, 3H, CH3); UV-Vis (e M−1cm−1): λ (nm) = 283 (15,000), 316 (8000).
- 3-amino-6,8-dibromo-2-methylquinazolin-4(3H)-one (3h) [60]: Yellow amorphous solid; mp: 231–232 °C; yield: 85%; 1H-NMR (DMSO-d6, 300 MHz): δ (ppm) 8.31 (d, J = 2.2 Hz, 1H, H7), 8.18 (d, J = 2.2 Hz, 1H, H5), 5.88 (s, 2H, NH2), 2.61 (s, 3H, CH3); UV-Vis (e M−1cm−1): λ (nm) = 286 (7285).
- 3-amino-6-iodo-2-methylquinazolin-4(3H)-one (3i) [61]: Off white amorphous solid; mp: 186–187 °C; yield: 63%; 1H-NMR (DMSO-d6, 600 MHz): δ (ppm) 8.36 (d, J = 2.1 Hz, 1H, H5), 8.04 (dd, J = 8.6, 2.1 Hz, 1H, H7), 7.39 (d, J = 8.6 Hz, 1H, H8), 5.82 (s, 2H, NH2), 2.56 (s, 3H, CH3); UV-Vis (e M−1cm−1): λ (nm) = 279 (13,253).
- 3-amino-2-methyl-6-nitroquinazolin-4(3H)-one (3j) [27]: Yellow amorphous solid; mp: 178–180 °C; yield: 85%; 1H-NMR (DMSO–d6, 600 MHz): δ (ppm) 8.80 (d, J = 2.3 Hz, 1H, H5), 8.49 (dd, J = 9.0, 2.4 Hz, 1H, H7), 7.78 (d, J = 8.9 Hz, 1H, H8), 5.92 (s, 2H, NH2), 2.64 (s, 3H, CH3); UV-Vis (e M−1cm−1): λ (nm) = 326 (9692).
- 3-amino-2-methyl-7-nitroquinazolin-4(3H)-one (3k) [62]: Yellow amorphous solid; mp: 218–219 °C; yield: 48%; 1H-NMR (DMSO-d6, 600 MHz): δ (ppm) 8.32 (m, 2H, H5, H8), 8.19 (dd, J = 8.64, 1.98 Hz, 1H, H6), 5.92 (s, 2H, NH2), 2.63 (s, 3H, CH3); UV-Vis (e M−1cm−1): λ (nm) = 258 (18,502), 344 (1762).
2.1.3. General Method of Synthesis of Acetamides (4–8)
- N-(2-methyl-4-oxoquinazolin-3(4H)-yl)acetamide (4) [63]: White amorphous solid; mp: 224–226 °C; yield: 87%; 1H NMR (CDCl3, 300 MHz): δ (ppm) 8.80 (s, 1H, NHCO), 8.19 (d, J = 7.8 Hz, 1H, H5), 7.76 (t, J = 8.1 Hz, 1H), 7.65 (d, J = 8.4Hz, 1H), 7.45 (t, J = 7.5 Hz, 1H), 2.54 (s, 3H, CH3), 2.26 (s, 3H, CH3); UV-Vis (e M−1cm−1): λ (nm) = 271 (12,200), 305 (7100).
- N-(6-chloro-2-methyl-4-oxoquinazolin-3(4H)-yl)acetamide (5): White amorphous solid; mp: 206–208 °C; yield: 45%; MS(ESI) m/z [M]+: 252; 1H NMR (DMSO-d6, 500 MHz): δ (ppm) 11.09 (s, 1H, NHCO), 8.04 (d, J = 2.4 Hz, 1H, H5), 7.88 (dd, J = 8.8, 2.5 Hz, 1H, H7), 7.67 (d, J = 8.7 Hz, 1H, H8), 2.38 (s, 3H, CH3), 2.11 (s, 3H, CH3); 13C-NMR (DMSO-d6, 125 MHz) δ 169.1, 158.0, 156.8, 145.3, 135.1, 130.9, 129.2, 125.3, 121.9, 21.1, 20.4; UV-Vis (e M−1cm−1): λ (nm) = 277 (14,300), 315 (6200).
- N-(6-bromo-2-methyl-4-oxoquinazolin-3(4H)-yl)acetamide (6) [63]: Off white amorphous solid; mp: 228–230 °C; yield: 49%; 1H NMR (DMSO-d6, 500 MHz): δ (ppm) 11.09 (s, 1H, NHCO), 8.17 (d, J = 2.3 Hz, 1H, H5), 7.99 (dd, J = 8.7, 2.3 Hz, 1H, H7), 7.59 (d, J = 8.7 Hz, 1H, H8), 2.37 (s, 3H, CH3), 2.11 (s, 3H, CH3); 13C-NMR (DMSO-d6, 125 MHz) δ 162.1, 157.8, 157.0, 145.6, 137.8, 129.3, 128.4, 122.2, 119.0, 21.1, 20.5; UV-Vis (e M−1cm−1): λ (nm) = 280 (15,200), 315 (7200).
- N-(6-iodo-2-methyl-4-oxoquinazolin-3(4H)-yl)acetamide (7): Beige amorphous solid; mp: 182–184 °C; yield: 42%; MS(ESI) m/z [M]+: 344; 1H NMR (DMSO-d6, 500 MHz): δ (ppm) 11.07 (s, 1H, NHCO), 8.35 (d, J = 2.0 Hz, 1H, H5), 8.12 (dd, J = 8.6, 2.0 Hz, 1H, H7), 7.42 (d, J = 8.6 Hz, 1H, H8), 2.36 (s, 3H, CH3), 2.10 (s, 3H, CH3); 13C-NMR (DMSO-d6, 125 MHz) δ 169.1, 157.7, 157.0, 145.9, 143.3, 134.5, 129.1, 122.4, 91.6, 21.1, 20.5; UV-Vis (e M−1cm−1): λ (nm) = 286 (15,800), 318 (6000).
- N-(2-methyl-6-nitro-4-oxoquinazolin-3(4H)-yl)acetamide (8): Yellow amorphous solid; mp: 242–244 °C; yield: 76%; MS(ESI) m/z [M]+: 263; 1H NMR (DMSO-d6, 500 MHz): δ (ppm) 11.21 (s, 1H, NHCO), 8.78 (d, J = 2.6 Hz, 1H, H5), 8.58 (dd, J = 9.0, 2.7 Hz, 1H, H7), 7.84 (d, J = 9.0 Hz, 1H, H8), 2.45 (s, 3H, CH3), 2.13 (s, 3H, CH3); 13C-NMR (DMSO-d6, 125 MHz) δ 169.2, 160.2, 158.2, 150.6, 145.0, 129.0, 128.8, 122.5, 120.7, 21.4, 20.4; UV-Vis (e M−1cm−1): λ (nm) = 270 (11,000), 318 (15,300), 329 (15,000).
2.1.4. General Method of Synthesis of Arylamides (9–19)
- 4-chloro-N-(2-methyl-4-oxoquinazolin-3(4H)-yl)benzamide (9) [64]: White amorphous solid; mp: 238–240 °C; yield: 72%; IR (KBr): 3234, 1704, 1667, 1606 cm−1; 1H-NMR (DMSO-d6, 500 MHz) δ 11.76 (brs, 1H, NHCO), 8.13 (d, J = 7.8 Hz, 1H), 8.02 (d, J = 8.4 Hz, 2H), 7.88 (t, J = 8.1 Hz, 1H), 7.69 (dd, J = 8.5, 2.1 Hz, 2H), 7.56 (t, J = 7.6 Hz, 1H), 2.46 (s, 3H, CH3) ppm; 13C-NMR (DMSO-d6, 125 MHz,) δ 165.0, 159.0, 156.1, 146.6, 137.7, 135.1, 130.0, 129.7, 129.0, 127.0, 126.9, 126.5, 120.6, 21.0 ppm; UV-Vis (e M−1cm−1): λ (nm) = 273 (3600), 305 (1600); HRMS(ESI) m/z [M+Na]+: C16H12ClN3NaO2+, calc: 336.0510; found: 336.0510.
- N-(2-methyl-4-oxoquinazolin-3(4H)-yl)-4-nitrobenzamide (10) [64]: Beige amorphous solid; mp: 269–272 °C; yield: 60%; IR (KBr): 3215, 1701, 1662, 1605 cm−1; 1H-NMR (DMSO-d6, 500 MHz) δ 12.04 (s, 1H, NHCO), 8.46 (d, J = 8.9 Hz, 2H), 8.25 (d, J = 8.9 Hz, 2H), 8.15 (dd, J = 7.9, 1.0 Hz, 1H), 7.91 (dt, J = 8.4, 1.4 Hz, 1H), 7.71 (d, J = 8 Hz, 1H), 7.58 (t, J = 7.9 Hz, 1H), 2.50 (s, 3H, CH3) ppm; 13C-NMR (DMSO-d6, 125 MHz,) δ 165.3, 159.4, 156.5, 150.4, 146.8, 137.1, 135.9, 129.8, 127.6, 127.3, 127.0, 124.5, 120.8, 21.4 ppm; UV-Vis (e M−1cm−1): λ (nm) = 275 (7040), broad declining shoulder up to ~340 nm; HRMS(ESI) m/z [M+H]+: C16H13N4O4+, calc: 325.0931; found: 325.0930.
- 4-methoxy-N-(2-methyl-4-oxoquinazolin-3(4H)-yl)benzamide (11) [65]: White amorphous solid; mp: 193–195 °C; yield: 82%; IR (KBr): 3215, 1702, 1659, 1608 cm−1; 1H-NMR (DMSO-d6, 500 MHz) δ 11.47 (s, 1H, NHCO), 8.12 (d, J = 7.8 Hz, 1H), 7.98 (d, J = 8.7 Hz, 2H), 7.88 (t, J = 8.1 Hz, 1H), 7.68 (d, J = 8.1 Hz, 1H), 7.56 (t, J = 7.4 Hz, 1H), 7.13 (d, J = 8.7 Hz, 2H), 3.87 (s, 3H, OCH3), 2.44 (s, 3H, CH3) ppm; 13C-NMR (DMSO-d6, 125 MHz,) δ 165.7, 163.0, 159.4, 156.7, 146.8, 135.4, 130.1, 127.1, 127.1, 126.7, 123.4, 120.8, 114.3, 55.8, 21.3 ppm; UV-Vis (e M−1cm−1): λ (nm) = 274 (18,000), 305 (5500); HRMS(ESI) m/z [M+Na]+: C17H15N3NaO3+, calc: 332.1006; found: 332.1003.
- N-(6-bromo-2-methyl-4-oxoquinazolin-3(4H)-yl)-4-chlorobenzamide (12): Off white amorphous solid; mp: 275–277 °C (EA+EtOH); yield: 76%; IR (KBr): 3263, 1698, 1668, 1603 cm−1; 1H-NMR (DMSO-d6, 500 MHz) δ 11.82 (s, 1H, NHCO), 8.21 (d, J = 2.1 Hz, 1H, H5), 8.04 (dd, J = 8.8, 2.2 Hz, 1H, H7), 8.01 (d, J = 8.5 Hz, 2H, ArCO), 7.69 (d, J = 8.5 Hz, 2H, ArCO), 7.65 (d, J = 8.7 Hz, 1H, H8), 2.45 (s, 3H, CH3) ppm; 13C-NMR (DMSO-d6, 125 MHz) δ 165.0, 157.9, 156.9, 145.6, 138.1, 137.8, 129.8, 129.8, 129.5, 129.0, 128.5, 122.2, 119.3, 21.1 ppm; UV-Vis (e M−1cm−1): λ (nm) = 274 (12,800), 315 (3800); HRMS(ESI) m/z [M+H]+: C16H12BrClN3O2+, calc: 391.9796; found: 391.9795.
- N-(6-bromo-2-methyl-4-oxoquinazolin-3(4H)-yl)-4-nitrobenzamide (13): Off white amorphous solid; yield: 65%; mp: 287–290 °C (EA+EtOH); IR (KBr): 3280, 1696, 1665, 1598 cm−1; 1H-NMR (DMSO-d6, 500 MHz) δ 12.09 (s, 1H, NHCO), 8.43 (d, J = 8.6 Hz, 2H, ArCO), 8.21 (d, J = 8.1 Hz, 2H, ArCO), 8.20 (obscured, 1H, H5), 8.03 (d, J = 8.8 Hz, 1H, H7), 7.64 (d, J = 8.7 Hz, 1H, H8), 2.47 (s, 3H, CH3) ppm; 13C-NMR (DMSO-d6, 125 MHz) δ 164.5, 157.8, 156.7, 150.0, 145.6, 138.1, 136.6, 129.5, 129.4, 128.5, 124.0, 122.1, 119.4, 21.1 ppm; UV-Vis (e M−1cm−1): λ (nm) = 280 (14,100), broad declining shoulder up to ~340 nm; HRMS(ESI) m/z [M+Na]+: C16H11BrN4NaO4+, calc: 424.9856; found: 424.9852.
- N-(6-bromo-2-methyl-4-oxoquinazolin-3(4H)-yl)-4-methoxybenzamide (14): White amorphous solid; yield: 78%; mp: 215–217 °C (EA); IR (KBr): 3311, 1700, 1664, 1605 cm−1; 1H-NMR (DMSO-d6, 500 MHz) δ 11.54 (s, 1H, NHCO), 8.21 (d, J = 2.1 Hz, 1H, H5), 8.03 (dd, J = 8.7, 2.1 Hz, 1H, H7), 7.98 (d, J = 8.7 Hz, 2H, ArCO), 7.65 (d, J = 8.7 Hz, 1H, H8), 7.13 (d, J = 8.7 Hz, 2H, ArCO), 3.86 (s, 3H, OCH3), 2.44 (s, 3H, CH3) ppm; 13C-NMR (DMSO-d6, 125 MHz) δ 165.3, 162.8, 158.0, 157.2, 145.6, 137.9, 129.9, 129.4, 128.4, 123.1, 122.2, 119.2, 114.1, 55.6, 21.1 ppm; UV-Vis (e M−1cm−1): λ (nm) = 278 (14,400), 315 (3800); HRMS(ESI) m/z [M+H]+: C17H15BrN3O3+, calc: 388.0291; found: 388.0287.
- N-(6-bromo-2-methyl-4-oxoquinazolin-3(4H)-yl)benzamide (15): Off white amorphous solid; yield: 84%; mp: 265–267 °C (EA); IR (KBr): 3262, 1694, 1669, 1607 cm−1; 1H-NMR (DMSO-d6, 500 MHz) δ 11.71 (s, 1H, NHCO), 8.22 (d, J = 2.2 Hz, 1H, H5), 8.04 (dd, J = 8.6, 2.2 Hz, 1H, H7), 8.00 (d, J = 7.3 Hz, 2H, ArCO), 7.70 (t, J = 7.5 Hz, 1H, ArCO), 7.65 (d, J = 8.7 Hz, 1H, H8), 7.61 (t, J = 7.6 Hz, 2H, ArCO), 2.46 (s, 3H, CH3) ppm; 13C-NMR (DMSO-d6, 125 MHz) δ 165.8, 157.9, 157.0, 145.6, 138.0, 132.9, 131.1, 129.4, 128.8, 128.4, 127.8, 122.2, 119.2, 21.1 ppm; UV-Vis (e M−1cm−1): λ (nm) = 274 (11,800), 315 (3200); HRMS(ESI) m/z [M+H]+: C16H13BrN3O2+, calc: 358.0186; found: 358.0178.
- 4-chloro-N-(2-methyl-6-nitro-4-oxoquinazolin-3(4H)-yl)benzamide (16): Pale yellow amorphous solid; yield: 58%; mp: 257–260 °C (EtOH); IR (KBr): 3273, 1705, 1681, 1601 cm−1; 1H-NMR (DMSO-d6, 500 MHz) δ 11.96 (s, 1H, NHCO), 8.82 (d, J = 2.6 Hz, 1H, H5), 8.61 (dd, J = 9.0, 2.6 Hz, 1H, H7), 8.03 (d, J = 8.5 Hz, 2H, ArCO), 7.90 (d, J = 9.0 Hz, 1H, H8), 7.70 (d, J = 8.5 Hz, 2H, ArCO), 2.54 (s, 3H, CH3) ppm; 13C-NMR (DMSO-d6, 125 MHz) δ 165.0, 160.1, 158.2, 150.6, 145.1, 137.8, 129.8, 129.7, 129.2, 129.0, 129.0, 122.6, 120.6, 21.5 ppm; UV-Vis (e M−1cm−1): λ (nm) = 265 (6800), 325 (10,800); HRMS(ESI) m/z [M+H]+: C16H12ClN4O4+, calc: 359.0542; found: 359.0549.
- N-(2-methyl-6-nitro-4-oxoquinazolin-3(4H)-yl)-4-nitrobenzamide (17): Beige amorphous solid; yield: 80%; mp: 244–246 °C (EtOH); IR (KBr): 3209, 1713, 1675, 1604 cm−1; 1H-NMR (DMSO-d6, 500 MHz) δ 12.24 (s, 1H, NHCO), 8.83 (d, J = 2.6 Hz, 1H, H5), 8.63 (dd, J = 9.0, 2.7 Hz, 1H, H7), 8.45 (d, J = 8.7 Hz, 2H, ArCO), 8.24 (d, J = 8.7 Hz, 2H, ArCO), 7.91 (d, J = 9.0 Hz, 1H, H8), 2.56 (s, 3H, CH3) ppm; 13C-NMR (DMSO-d6, 125 MHz) δ 164.5, 159.9, 158.0, 150.6, 150.0, 145.2, 136.5, 129.5, 129.3, 129.0, 124.0, 122.6, 120.6, 21.4 ppm; UV-Vis (e M−1cm−1): λ (nm) = 270 (12,700), 321 (12,900); HRMS(ESI) m/z [M+Na]+: C16H11N5NaO6+, calc: 392.0602; found: 392.0604.
- 4-methoxy-N-(2-methyl-6-nitro-4-oxoquinazolin-3(4H)-yl)benzamide (18): Beige amorphous solid; yield: 84%; mp: 228–230 °C (EtOH); IR (KBr): 3306, 1699, 1678, 1606 cm−1; 1H-NMR (DMSO-d6, 500 MHz) δ 11.67 (s, 1H, NHCO), 8.81 (d, J = 2.6 Hz, 1H, H5), 8.61 (dd, J = 8.9, 2.6 Hz, 1H, H7), 8.00 (d, J = 8.8 Hz, 2H, ArCO), 7.90 (d, J = 9.0 Hz, 1H, H8), 7.14 (d, J = 8.7 Hz, 2H, ArCO), 3.87 (s, 3H, OCH3), 2.52 (s, 3H, CH3) ppm; 13C-NMR (DMSO-d6, 125 MHz) δ 165.4, 163.0, 160.4, 158.4, 150.7, 145.1, 130.0, 129.2, 129.0, 123.0, 122.7, 120.7, 114.2, 55.6, 21.5 ppm; UV-Vis (e M−1cm−1): λ (nm) = 269 (11,200), 326 (10,900); HRMS(ESI) m/z [M+Na]+: C17H14N4NaO5+, calc: 377.0856; found: 377.0856.
- N-(2-methyl-6-nitro-4-oxoquinazolin-3(4H)-yl)benzamide (19): Off white amorphous solid; yield: 87%; mp: 262–264 °C (EtOH); IR (KBr): 3271, 1690, 1678, 1603 cm−1; 1H-NMR (DMSO-d6, 500 MHz) δ 11.85 (s, 1H, NHCO), 8.82 (d, J = 2.6 Hz, 1H, H5), 8.62 (dd, J = 8.9, 2.7 Hz, 1H, H7), 8.01 (d, J = 7.2 Hz, 2H, ArCO), 7.90 (d, J = 9.0 Hz, 1H, H8), 7.71 (t, J = 7.5 Hz, 1H, ArCO), 7.62 (t, J = 7.7 Hz, 2H, ArCO), 2.54 (s, 3H, CH3) ppm, 13C-NMR (DMSO-d6, 125 MHz) δ 165.9, 160.2, 158.3, 150.6, 145.1, 133.0, 131.0, 129.2, 129.0, 128.9, 127.9, 122.6, 120.7, 21.5 ppm; UV-Vis (e M−1cm−1): λ (nm) = 265 (5800), 325 (11,500); HRMS(ESI) m/z [M+H]+: C16H13N4O4+, calc: 325.0931; found: 325.0932.
2.2. Supercoiled Circular pB322 DNA Photocleavage Experiments by QNZs 3a-k, 4-8, 11-19
2.3. Molecular Docking Studies
3. Results and Discussion
3.1. Synthesis
3.2. DNA Photocleavage
3.3. DNA Molecular Docking Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- He, D.; Wang, M.; Zhao, S.; Shu, Y.; Zeng, H.; Xiao, C.; Lu, C.; Liu, Y. Pharmaceutical prospects of naturally occurring quinazolinone and its derivatives. Fitoterapia 2017, 119, 136–149. [Google Scholar] [CrossRef]
- Mhaske, S.B.; Argade, N.P. The chemistry of recently isolated naturally occurring quinazolinone alkaloids. Tetrahedron 2006, 62, 9787–9826. [Google Scholar] [CrossRef]
- Khan, I.; Zaib, S.; Batool, S.; Abbas, N.; Ashraf, Z.; Iqbal, J.; Saeed, A. Quinazolines and quinazolinones as ubiquitous structural fragments in medicinal chemistry: An update on the development of synthetic methods and pharmacological diversification. Bioorg. Med. Chem. 2016, 24, 2361–2381. [Google Scholar] [CrossRef] [PubMed]
- Auti, P.S.; George, G.; Paul, A.T. Recent advances in the pharmacological diversification of quinazoline/quinazolinone hybrids. RSC Adv. 2020, 10, 41353–41392. [Google Scholar] [CrossRef]
- Mohammadkhani, L.; Heravi, M.M. Microwave-Assisted Synthesis of Quinazolines and Quinazolinones: An Overview. Front. Chem. 2020, 8, 580086. [Google Scholar] [CrossRef] [PubMed]
- Pathak, P.; Shukla, P.K.; Kumar, V.; Kumar, A.; Verma, A. Quinazoline clubbed 1,3,5-triazine derivatives as VEGFR2 kinase inhibitors: Design, synthesis, docking, in vitro cytotoxicity and in ovo antiangiogenic activity. Inflammopharmacology 2018, 26, 1441–1453. [Google Scholar] [CrossRef]
- Sahu, S.K.; Azam, A.; Banerjee, M.; Acharrya, S.; Behera, C.C.; Si, S. Synthesis, characterization and biological activity of 2-methyl-3-aminoquinazolin-4(3H)-ones Schiff bases. J. Braz. Chem. Soc. 2008, 19, 963–970. [Google Scholar] [CrossRef]
- Gao, X.; Cai, X.; Yan, K.; Song, B.; Gao, L.; Chen, Z. Synthesis and Antiviral Bioactivities of 2-Aryl- or 2-Methyl-3-(substituted- Benzalamino)-4(3H)-quinazolinone Derivatives. Molecules 2007, 12, 2621–2642. [Google Scholar] [CrossRef] [Green Version]
- Jafari, E.; Khajouei, M.R.; Hassanzadeh, F.; Hakimelahi, G.H.; Khodarahmi, G.A. Quinazolinone and quinazoline derivatives: Recent structures with potent antimicrobial and cytotoxic activities. Res. Pharm. Sci. 2016, 11, 1–14. [Google Scholar]
- Sawant, S.D.; Sahu, M.; Nerkar, A.G. Quinazolinone Platinum Metal Complexes: In silico Design, Synthesis and Evaluation of Anticancer Activity. Asian J. Chem. 2018, 30, 2164–2170. [Google Scholar] [CrossRef]
- El-Sayed, N.N.E.; Almaneai, N.M.; Ben Bacha, A.; Al-Obeed, O.; Ahmad, R.; Abdulla, M.; Alafeefy, A.M. Synthesis and evaluation of anticancer, antiphospholipases, antiproteases, and antimetabolic syndrome activities of some 3H-quinazolin-4-one derivatives. J. Enzym. Inhib. Med. Chem. 2019, 34, 672–683. [Google Scholar] [CrossRef] [Green Version]
- Su, C.; Liu, X. Synthesis of Novel 4(3H)-Quinazolinones with 1,2,3-Triazoles Moiety Conjugated by Schiff Base. Asian J. Chem. 2014, 26, 5301–5304. [Google Scholar] [CrossRef]
- Raghavendra, N.M.; Thampi, P.; Gurubasavarajaswamy, P.M.; Sriram, D. Synthesis, Antitubercular and Anticancer Activities of Substituted Furyl-quinazolin-3(4H)-ones. Arch. Pharm. 2007, 340, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Raghavendra, N.M.; Thampi, P.; Gurubasavarajaswamy, P.M.; Sriram, D. Synthesis and Antimicrobial Activities of Some Novel Substituted 2-Imidazolyl-N-(4-oxo-quinazolin-3(4H)-yl)-acetamides. Chem. Pharm. Bull. 2007, 55, 1615–1619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, Z.; Wu, W.; Miao, Y.; Tang, Y.; Zhou, Y.; Zheng, L.; Fu, Y.; Song, Z.; Peng, Y. Recent advances in quinazolinones as an emerging molecular platform for luminescent materials and bioimaging. Org. Chem. Front. 2021, 8, 1867–1889. [Google Scholar] [CrossRef]
- Bajaj, K.; Srivastava, V.K.; Kumar, A. Newer Substituted Benzoxazepinylquinazolinones as Potent Antipsychotic and Anticonvulsant Agents. Arzneimittelforschung 2003, 53, 480–485. [Google Scholar] [CrossRef]
- Kashaw, S.K.; Kashaw, V.; Mishra, P.; Jain, N.K.; Stables, J.P. Design, synthesis, and potential CNS activity of some new bioactive 1-(4-substituted-phenyl)-3-(4-oxo-2-methyl-4H-quinazolin-3-yl)-urea. Med. Chem. Res. 2011, 20, 738–745. [Google Scholar] [CrossRef]
- Alagarsamy, V.; Murugananthan, G.; Venkateshperumal, R. Synthesis, Analgesic, Anti-inflammatory and Antibacterial Activities of Some Novel 2-Methyl-3-substituted Quinazolin-4-(3H)-ones. Biol. Pharm. Bull. 2003, 26, 1711–1714. [Google Scholar] [CrossRef] [Green Version]
- Ghabrial, S.S.; Gaber, H.M. Dipolar Cycloaddition Reactions with Quinazolinones: A New Route for the Synthesis of Several Annelated Pyrrolo- and Pyridazinoquinazoline Derivatives. Molecules 2003, 8, 401–410. [Google Scholar] [CrossRef] [Green Version]
- Zeydi, M.M.; Montazeri, N.; Fouladi, M. Synthesis and Evaluation of Novel [1,2,4]Triazolo[1,5-c]quinazoline Derivatives as Antibacterial Agents. J. Heterocycl. Chem. 2017, 54, 3549–3553. [Google Scholar] [CrossRef]
- Farag, D.B.; Farag, N.A.; Esmat, A.; Abuelezz, S.A.; Ibrahim, E.A.-S.; El Ella, D.A.A. Synthesis, 3D pharmacophore, QSAR and docking studies of novel quinazoline derivatives with nitric oxide release moiety as preferential COX-2 inhibitors. MedChemComm 2015, 6, 283–299. [Google Scholar] [CrossRef]
- Dinari, M.; Gharahi, F.; Asadi, P. Synthesis, spectroscopic characterization, antimicrobial evaluation and molecular docking study of novel triazine-quinazolinone based hybrids. J. Mol. Struct. 2018, 1156, 43–50. [Google Scholar] [CrossRef]
- Hassanzadeh, F.; Sadeghi-Aliabadi, H.; Jafari, E.; Sharifzadeh, A.; Dana, N. Synthesis and cytotoxic evaluation of some quinazolinone- 5-(4-chlorophenyl) 1, 3, 4-oxadiazole conjugates. Res. Pharm. Sci. 2019, 14, 408–413. [Google Scholar] [CrossRef] [PubMed]
- Fleita, D.H.; Mohareb, R.M.; Sakka, O.K. Antitumor and antileishmanial evaluation of novel heterocycles derived from quinazoline scaffold: A molecular modeling approach. Med. Chem. Res. 2013, 22, 2207–2221. [Google Scholar] [CrossRef]
- Babu, R.R.; Naresh, K.; Ravi, A.; Reddy, B.M.; Babu, V.H. Synthesis of novel isoniazid incorporated styryl quinazolinones as anti-tubercular agents against INH sensitive and MDR M. tuberculosis strains. Med. Chem. Res. 2014, 23, 4414–4419. [Google Scholar] [CrossRef]
- Ponomarev, I.I.; Razorenov, D.Y.; Petrovskii, P.V. Synthesis of poly(pyridazinoquinazolones). Russ. Chem. Bull. 2009, 58, 2376–2384. [Google Scholar] [CrossRef]
- Patel, V.H.; Patel, M.P.; Patel, R.G. Synthesis and Application of Novel Heterocyclic Dyes Based on 11-Amino-3-Bromo-13h-Acenaphtho[1,2-E]Pyridazino[3,2-B]- Quinazoli Ne-13-One. Heterocycl. Commun. 2001, 7, 599–606. [Google Scholar] [CrossRef]
- Maiti, S.; Kim, J.; Park, J.-H.; Nam, D.; Lee, J.B.; Kim, Y.-J.; Kee, J.-M.; Seo, J.K.; Myung, K.; Rohde, J.-U.; et al. Chemoselective Trifluoroethylation Reactions of Quinazolinones and Identification of Photostability. J. Org. Chem. 2019, 84, 6737–6751. [Google Scholar] [CrossRef]
- Fan, W.; Huang, P.; Chen, X. Overcoming the Achilles’ heel of photodynamic therapy. Chem. Soc. Rev. 2016, 45, 6488–6519. [Google Scholar] [CrossRef]
- Zhang, J.; Jiang, C.; Figueiró Longo, J.P.; Azevedo, R.B.; Zhang, H.; Muehlmann, L.A. An updated overview on the development of new photosensitizers for anticancer photodynamic therapy. Acta Pharm. Sin. B 2018, 8, 137–146. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Wang, Y.; Murray, C.K.; Hamblin, M.R.; Hooper, D.C.; Dai, T. Antimicrobial blue light inactivation of pathogenic microbes: State of the art. Drug Resist. Updat. 2017, 33–35, 1–22. [Google Scholar] [CrossRef]
- Shleeva, M.; Savitsky, A.; Kaprelyants, A. Photoinactivation of mycobacteria to combat infection diseases: Current state and perspectives. Appl. Microbiol. Biotechnol. 2021, 105, 4099–4109. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, S.K.; Singh, H.; Deep, A.; Khatri, M.; Bhaumik, J.; Kim, K.-H.; Bhardwaj, N. UVC-based photoinactivation as an efficient tool to control the transmission of coronaviruses. Sci. Total Environ. 2021, 792, 148548. [Google Scholar] [CrossRef] [PubMed]
- Sobotta, L.; Skupin-Mrugalska, P.; Piskorz, J.; Mielcarek, J. Non-porphyrinoid photosensitizers mediated photodynamic inactivation against bacteria. Dye. Pigment. 2019, 163, 337–355. [Google Scholar] [CrossRef]
- Hamblin, M.R.; Abrahamse, H. Oxygen-Independent Antimicrobial Photoinactivation: Type III Photochemical Mechanism? Antibiotics 2020, 9, 53. [Google Scholar] [CrossRef] [Green Version]
- Ovchinnikova, I.G.; Kim, G.A.; Matochkina, E.G.; Kodess, M.I.; Slepukhin, P.A.; Kovalev, I.S.; Nosova, E.V.; Rusinov, G.L.; Charushin, V.N. Synthesis, photochemical and luminescent properties of ortho-hydroxystyrylquinazolinone-linked benzocrown ethers. J. Photochem. Photobiol. A Chem. 2018, 351, 16–28. [Google Scholar] [CrossRef]
- Kim, G.; Ovchinnikova, I.G.; Nosova, E.V.; Rusinov, G.L.; Charushin, V.N. (E)-2-(Hydroxystyryl)-3-phenylquinazolin-4(3H)-ones: Synthesis, photochemical and luminescent properties. Arkivoc 2018, 2018, 266–277. [Google Scholar] [CrossRef] [Green Version]
- Hricovíni, M.; Hricovíni, M. Photochemically-induced anti-syn isomerization of quinazolinone-derived Schiff’s bases: EPR, NMR and DFT analysis. Tetrahedron 2017, 73, 252–261. [Google Scholar] [CrossRef]
- Tokura, Y.; Ogai, M.; Yagi, H.; Takigawa, M. Afloqualone Photosensitivity. Immunogenicity of Afloqualone-Photomodified Epidermal Cells. Photochem. Photobiol. 1994, 60, 262–267. [Google Scholar] [CrossRef]
- Fujita, H.; Matsuo, I. Phototoxic potential of afloqualone, a quinazolinone derivative, as determined by photosensitized inactivation of bacteriophage. Chem. Biol. Interact. 1987, 64, 139–149. [Google Scholar] [CrossRef]
- Fujita, H.; Matsuo, I. UV-A induced DNA nicking activities of skin photosensitive drugs: Phenothiazines, benzothiadiazines and afloqualone. Chem. Biol. Interact. 1988, 66, 27–36. [Google Scholar] [CrossRef]
- Iwamoto, Y.; Itoyama, T.; Yasuda, K.; Uzuhashi, T.; Tanizawa, H.; Takino, Y.; Oku, T.; Hashizume, H.; Yanagihara, Y. Photodynamic Deoxyribonucleic Acid (DNA) Strand Breaking Activities of Enoxacin and Afloqualone. Chem. Pharm. Bull. 1992, 40, 1868–1870. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Liu, L.; Pan, Y.; Zhu, Q.; Lu, Y.; Wei, J.; Luo, K.; Fu, Y.; Zhong, C.; Peng, Y.; et al. Asymmetric Difluoroboron Quinazolinone-Pyridine Dyes with Large Stokes Shift: High Emission Efficiencies Both in Solution and in the Solid State. Chem. A Eur. J. 2018, 24, 17897–17901. [Google Scholar] [CrossRef]
- Lazou, M.; Tarushi, A.; Gritzapis, P.; Psomas, G. Transition metal complexes with a novel guanine-based (E)-2-(2-(pyridin-2-ylmethylene)hydrazinyl)quinazolin-4(3H)-one: Synthesis, characterization, interaction with DNA and albumins and antioxidant activity. J. Inorg. Biochem. 2020, 206, 111019. [Google Scholar] [CrossRef]
- Kakoulidou, C.; Gritzapis, P.S.; Hatzidimitriou, A.G.; Fylaktakidou, K.C.; Psomas, G. Zn(II) complexes of (E)-4-(2-(pyridin-2-ylmethylene)hydrazinyl)quinazoline in combination with non-steroidal anti-inflammatory drug sodium diclofenac: Structure, DNA binding and photo-cleavage studies, antioxidant activity and interaction with albumin. J. Inorg. Biochem. 2020, 211, 111194. [Google Scholar] [CrossRef]
- Kakoulidou, C.; Kosmas, V.-R.; Hatzidimitriou, A.G.; Fylaktakidou, K.C.; Psomas, G. Structure and biological profile of transition metal complexes with (E)-4-(2-(pyridin-2-ylmethylene)hydrazinyl)quinazoline. J. Inorg. Biochem. 2021, 219, 111448. [Google Scholar] [CrossRef]
- Kakoulidou, C.; Hatzidimitriou, A.G.; Fylaktakidou, K.C.; Psomas, G. Interaction of manganese(II) with the hybrid molecule (E)-4-(2-(pyridin-2-ylmethylene)hydrazinyl)quinazoline: Structure and biological profile. Polyhedron 2021, 195, 114986. [Google Scholar] [CrossRef]
- Peng, X.-M.; Peng, L.-P.; Li, S.; Avula, S.R.; Kannekanti, V.K.; Zhang, S.-L.; Tam, K.Y.; Zhou, C.-H. Quinazolinone azolyl ethanols: Potential lead antimicrobial agents with dual action modes targeting methicillin-resistant Staphylococcus aureus DNA. Future Med. Chem. 2016, 8, 1927–1940. [Google Scholar] [CrossRef]
- Peng, L.-P.; Nagarajan, S.; Rasheed, S.; Zhou, C.-H. Synthesis and biological evaluation of a new class of quinazolinoneazoles as potential antimicrobial agents and their interactions with calf thymus DNA and human serum albumin. MedChemComm 2015, 6, 222–229. [Google Scholar] [CrossRef]
- Panagopoulos, A.; Balalas, T.; Mitrakas, A.; Vrazas, V.; Katsani, K.R.; Koumbis, A.E.; Koukourakis, M.I.; Litinas, K.E.; Fylaktakidou, K.C. 6-Nitro-Quinazolin−4(3H)−one Exhibits Photodynamic Effects and Photodegrades Human Melanoma Cell Lines. A Study on the Photoreactivity of Simple Quinazolin−4(3H)−ones. Photochem. Photobiol. 2021, 97, 826–836. [Google Scholar] [CrossRef]
- Armitage, B. Photocleavage of Nucleic Acids. Chem. Rev. 1998, 98, 1171–1200. [Google Scholar] [CrossRef]
- Cadet, J.; Mouret, S.; Ravanat, J.-L.; Douki, T. Photoinduced Damage to Cellular DNA: Direct and Photosensitized Reactions. Photochem. Photobiol. 2012, 88, 1048–1065. [Google Scholar] [CrossRef]
- Pasolli, M.; Dafnopoulos, K.; Andreou, N.-P.; Gritzapis, P.S.; Koffa, M.; Koumbis, A.E.; Psomas, G.; Fylaktakidou, K.C. Pyridine and p-Nitrophenyl Oxime Esters with Possible Photochemotherapeutic Activity: Synthesis, DNA Photocleavage and DNA Binding Studies. Molecules 2016, 21, 864. [Google Scholar] [CrossRef] [Green Version]
- Gritzapis, P.S.; Varras, P.C.; Andreou, N.-P.; Katsani, K.R.; Dafnopoulos, K.; Psomas, G.; Peitsinis, Z.V.; Koumbis, A.E.; Fylaktakidou, K.C. p-Pyridinyl oxime carbamates: Synthesis, DNA binding, DNA photocleaving activity and theoretical photodegradation studies. Beilstein J. Org. Chem. 2020, 16, 337–350. [Google Scholar] [CrossRef] [Green Version]
- Peet, N.P.; Sunder, S. Factors Which Influence the Formation of Oxadiazoles from Anthranilhydrazides and other Benoylhydrazines. J. Heterocycl. Chem. 1984, 21, 1807–1816. [Google Scholar] [CrossRef]
- Gavin, J.; Annor-Gyamfi, J.K.; Bunce, R.A. Quinazolin-4(3H)-ones and 5,6-Dihydropyrimidin-4(3H)-ones from β-Aminoamides and Orthoesters. Molecules 2018, 23, 2925. [Google Scholar] [CrossRef] [Green Version]
- Lambeth, J.D.; Ganesh, T.; Smith, S.M. Quinazoline Derivatives, Compositions, and Uses Related Thereto. WIPO Patent WO2012058211A2, 3 May 2012. [Google Scholar]
- Vlaovic, D.; Milic, B.L.; Mackenzie, K. Modified procedure for the preparation of 5-nitro-2-furylmethylene diacetate and its use in the synthesis of some novel (5-nitro-2-furyl) azomethines via 5-nitro-2-furaldehyde. J. Chem. Res.-S 1989, 20. [Google Scholar] [CrossRef]
- Wei, W.; Zhu, L.; Zhou, Y.; Li, Z. Synthesis and Bioactivities Evaluation of Novel N-Pyridylpyrazole Derivatives with 1,2,3-Triazole and Quinazolin-4(3H)-one Substructures. Heterocycles 2018, 96, 1453–1462. [Google Scholar] [CrossRef]
- ElZahabi, H.S.; Nafie, M.S.; Osman, D.; Elghazawy, N.H.; Soliman, D.H.; El-Helby, A.A.H.; Arafa, R.K. Design, synthesis and evaluation of new quinazolin-4-one derivatives as apoptotic enhancers and autophagy inhibitors with potent antitumor activity. Eur. J. Med. Chem. 2021, 222, 113609. [Google Scholar] [CrossRef]
- Babu, Y.R.; Bhagavanraju, M.; Reddy, G.D.; Peters, G.J.; Prasad, V.V.S.R. Design and Synthesis of Quinazolinone Tagged Acridones as Cytotoxic Agents and Their Effects on EGFR Tyrosine Kinase. Arch. Pharm. 2014, 347, 624–634. [Google Scholar] [CrossRef]
- Buyuktimkin, S. Quinazolinones. Part 4: Synthesis of 2-methyl-3-(4-oxo-thiazolidin-2-ylidenanimo)-4(3H)-quinazolinone derivatives | Chinazolinone. 4. Mitteilung: Synthese von Derivaten des 2-Methyl-3-(4-Oxo-Thiazolidin-2-Ylidenamino)-4(3h)-Chinazolinons. Pharmazie 1985, 40, 393–395. [Google Scholar]
- Archana, A.; Srivastava, V.K.; Chandra, R.; Kumar, A. Synthesis of potential quinazolinonyl pyrazolines and quinazolinyl isoxazolines as anticonvulsant agents. Indian J. Chem. 2002, 41, 2371–2375. [Google Scholar] [CrossRef]
- Varnavas, A.; Lassiani, L.; Luxich, E.; Zacchigna, M.; Boccù, E. Quinazolinone derivatives: Synthesis and binding evaluation on cholecystokinin receptors. Farmarco 1996, 51, 333–339. [Google Scholar]
- Ibrahim, S.S.; Abdel-Halim, A.M.; Gabr, Y.; El-Edfawy, S.; Abdel-Rahman, R.M. Synthesis and Biological Evaluation of Some New Fused Quinazoline Derivatives. J. Chem. Res. 1997, 27, 154–155. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision B.01; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Drew, H.R.; Wing, R.M.; Takano, T.; Broka, C.; Tanaka, S.; Itakura, K.; Dickerson, R.E. Structure of a B-DNA dodecamer: Conformation and dynamics. Proc. Natl. Acad. Sci. USA 1981, 78, 2179–2183. [Google Scholar] [CrossRef] [Green Version]
- Trott, O.; Olson, A.J. Software news and update AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar]
- DeLano, W.L. The PyMOL Molecular Graphics System; Delano Scientific: San Carlos, CA, USA, 2002. [Google Scholar]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. Des. Sel. 1995, 8, 127–134. [Google Scholar] [CrossRef]
- Tobiszewski, M.; Namieśnik, J.; Pena-Pereira, F. Environmental risk-based ranking of solvents using the combination of a multimedia model and multi-criteria decision analysis. Green Chem. 2017, 19, 1034–1042. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.; Chen, D.; Chou, P. Fluorescent Chromophores Containing the Nitro Group: Relatively Unexplored Emissive Properties. ChemPlusChem 2021, 86, 11–27. [Google Scholar] [CrossRef]
- Scholz, M.; Dědic, R.; Svoboda, A.; Hála, J. TPP and singlet oxygen quenching by carotene in solution. J. Mol. Struct. 2011, 993, 474–476. [Google Scholar] [CrossRef]
- Zobel, J.P.; Nogueira, J.J.; González, L. Mechanism of Ultrafast Intersystem Crossing in 2-Nitronaphthalene. Chem. A Eur. J. 2018, 24, 5379–5387. [Google Scholar] [CrossRef] [Green Version]










| A/A | No | Compound Text Representation a | E Binding (kcal/mol) | Polar Contacts (PyMOL) | Interactions (LigPlus) |
|---|---|---|---|---|---|
| Group A (6-H) | 3c | 6-H-QNZ-3-NH2 | −6.7 | DG16, DC15, DG10, DC11 | nc b |
| 4 | 6-H-QNZ-3-NHCOMe | −7.1 | DG16, DG10, DC11 | nc b | |
| 9 | 6-H-QNZ-3-NHCOAr-Cl | −9.0 | DG16, DG10 | nc b | |
| 10 | 6-H-QNZ-3-NHCOAr-NO2 | −9.4 | DA5, DG4, DG22 | HB: DG4(A) (4−C=O) | |
| VdW: DA5(A) (4−C=O), DA6(A), DC21(B), DG22(B), DC23(B) | |||||
| 11 | 6-H-QNZ-3-NHCOAr-OMe | −9.3 | DG16, DG10 | nc b | |
| - | 6-H-QNZ-3-NHCOAr-H | −8.8 | DG4, DA5, DG22 | nc b |
| A/A | No | Compound Text Representation a | E Binding (kcal/mol) | Polar Contacts (PyMOL) | Interactions (LigPlus) |
|---|---|---|---|---|---|
| Group B (6−Br) | 3g | 6-Br-QNZ-3-NH2 | −7.0 | DG16, DC15, DG10, DC11 | HB: DG10(A) (4−C=O, 3−NH2), DG16(B) (4−C=O), DC15(B) (3−NH2) |
| VdW: DC11(A), DC17(B), DG12(A) | |||||
| 6 | 6-Br-QNZ-3-NHCOMe | −7.8 | DC3, DG4, DA5, DG22 | HB: DG4(A) (4−C=O), DG22(B) (Me−C=O) | |
| VdW: DA5(A) (4−C=O), DC23(B), DA6(A), DT7(A) (Br), DC21(B) | |||||
| 12 | 6-Br-QNZ-3-NHCOAr-Cl | −9.2 | DG10, DC11 | ncb | |
| 13 | 6-Br-QNZ-3-NHCOAr-NO2 | −9.6 | DG10, DC11 | HB: DG10(A) (4−C=O) | |
| VdW: DA18(B), DA17(B), DC11(A), DG12(A), DG16(B), DG14(B), DC15(B) (Ar−NO−O) | |||||
| 14 | 6-Br-QNZ-3-NHCOAr-OMe | −9.4 | DG16, DC9 | nc b | |
| 15 | 6-Br-QNZ-3-NHCOAr-H | −8.9 | DG10, DC11 | nc b |
| A/A | No | Compound Text Representation a | E Binding (kcal/mol) | Polar Contacts b (PyMOL) | Interactions (LigPlus) |
|---|---|---|---|---|---|
| Group C (6-NO2) | 3j | 6-NO2-QNZ-3-NH2 | −7.6 | DG16, DC15, DG10, DC11 | HB: DG10(A) (4−C=O, 3−NH2), DG16(B) (4−C=O), DC15(B) (3−NH2), DC11(A) (3−NH2) |
| VdW: DG12(A), DA17(B), DA18(B), [DC21(B), DT7(A), DA6(A) (6−NO−O)], (6−NO−O) | |||||
| 8 | 6-NO2-QNZ-3-NHCOMe | −8.8 | DG4, DA5, DA6, DG22 | HB: DG4(A) (4−C=O) | |
| VdW: DC23(A) (Me−C=O), DG22(B), [DC21(B), DT7(A), DA6(A) (6−NO−O)], DA5(A) (4−C=O) | |||||
| 16 | 6-NO2-QNZ-3-NHCOAr-Cl | −9.8 | DG16, DG10, DC11 | HB: DG16(B), DG10(A) | |
| VdW: DC11(A), DG12(A), DG14(B), DC15(B), DA17(B), DA18(B) | |||||
| 17 | 6-NO2-QNZ-3-NHCOAr-NO2 | −10.1 | DG2, DG4, DG22 | HB: DG2(A) (6−NO−O), [DG4(A), DG22(B) (4−C=O)] | |
| VdW: DC3(A) (6−NO−O), DA5(A), DA6(A), DC21(B) (6−NO−O) | |||||
| 18 | 6-NO2-QNZ-3-NHCOAr-OMe | −10.0 | DG16, DG10, DC11 | nc b | |
| 19 | 6-NO2-QNZ-3-NHCOAr-H | −9.5 | DG16, DG10, DC11 | nc b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mikra, C.; Bairaktari, M.; Petridi, M.-T.; Detsi, A.; Fylaktakidou, K.C. Green Process for the Synthesis of 3-Amino-2-methyl-quinazolin-4(3H)-one Synthones and Amides Thereof:DNA Photo-Disruptive and Molecular Docking Studies. Processes 2022, 10, 384. https://0-doi-org.brum.beds.ac.uk/10.3390/pr10020384
Mikra C, Bairaktari M, Petridi M-T, Detsi A, Fylaktakidou KC. Green Process for the Synthesis of 3-Amino-2-methyl-quinazolin-4(3H)-one Synthones and Amides Thereof:DNA Photo-Disruptive and Molecular Docking Studies. Processes. 2022; 10(2):384. https://0-doi-org.brum.beds.ac.uk/10.3390/pr10020384
Chicago/Turabian StyleMikra, Chrysoula, Maria Bairaktari, Marina-Theodora Petridi, Anastasia Detsi, and Konstantina C. Fylaktakidou. 2022. "Green Process for the Synthesis of 3-Amino-2-methyl-quinazolin-4(3H)-one Synthones and Amides Thereof:DNA Photo-Disruptive and Molecular Docking Studies" Processes 10, no. 2: 384. https://0-doi-org.brum.beds.ac.uk/10.3390/pr10020384