Prevalence, Antibiogram and Genetic Characterization of Listeria monocytogenes from Food Products in Egypt
Abstract
:1. Introduction
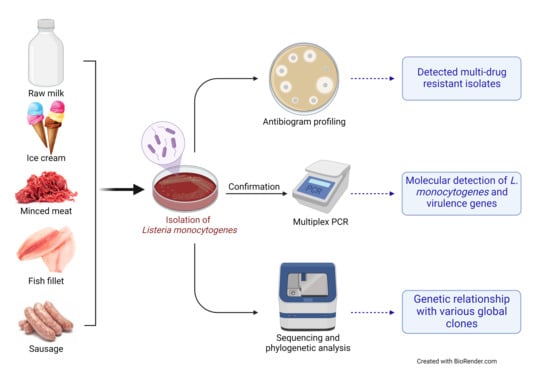
2. Materials and Methods
2.1. Ethics Statement
2.2. Study Area
2.3. Samples Collection and Processing
2.4. Phenotypic Isolation and Identification of Listeria from Food Products
2.5. Antibiogram Profile of L. monocytogenes Isolates Recovered from Food Products
2.6. Molecular Detection of L. monocytogenes from Food Products
2.7. Sequencing and Phylogenetic Analysis of L. monocytogenes from Food Products
2.8. Statistical Analysis
3. Results
3.1. Prevalence of Listeria Species and L. monocytogenes in Different Food Products
3.2. Antibiogram of L. monocytogenes Isolates from Food Products
3.3. Screening of Different Virulence Genes in L. monocytogenes from Food Products
3.4. Sequence Analysis of L. monocytogenes Isolates from Food Products
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Osman, K.M.; Kappell, A.D.; Fox, E.M.; Orabi, A.; Samir, A. Prevalence, pathogenicity, virulence, antibiotic resistance, and phylogenetic analysis of biofilm producing Listeria monocytogenes isolated from different ecological niches in Egypt: Food, humans, animals, and environment. Pathogens 2020, 9, 5. [Google Scholar] [CrossRef] [Green Version]
- Skowron, K.; Wałecka-Zacharksa, E.; Grudlewska, K.; Wiktorczyk, N.; Kaczmarek, A.; Gryń, G.; Kwiecińska-Piróg, J.; Juszczuk, K.; Paluszak, Z.; Kosek-Paszkowska, K.; et al. Characteristics of Listeria monocytogenes strains isolated from milk and humans and the possibility of milk-borne strains transmission. Polish J. Microbiol. 2019, 68, 353–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinothkumar, R.; Arunagiri, K.; Sivakumar, T. Studies on pathogenic Listeria monocytogenes from marine food resources. Int. J. Curr. Microbiol. Appl. Sci. 2013, 2, 86–93. [Google Scholar]
- Kim, S.W.; Haendiges, J.; Keller, E.N.; Myers, R.; Kim, A.; Lombard, J.E.; Karns, J.S.; Van Kessel, J.A.S.; Haley, B.J. Genetic diversity and virulence profiles of Listeria monocytogenes recovered from bulk tank milk, milk filters, and milking equipment from dairies in the United States (2002 to 2014). PLoS ONE 2018, 13, e0197053. [Google Scholar] [CrossRef] [PubMed]
- Maury, M.M.; Bracq-Dieye, H.; Huang, L.; Vales, G.; Lavina, M.; Thouvenot, P.; Disson, O.; Leclercq, A.; Brisse, S.; Lecuit, M. Hypervirulent Listeria monocytogenes clones’ adaption to mammalian gut accounts for their association with dairy products. Nat. Commun. 2019, 10, 2488. [Google Scholar] [CrossRef] [Green Version]
- Camejo, A.; Carvalho, F.; Reis, O.; Leitão, E.; Sousa, S.; Cabanes, D. The arsenal of virulence factors deployed by Listeria monocytogenes to promote its cell infection cycle. Virulence 2011, 2, 379–394. [Google Scholar] [CrossRef] [Green Version]
- Ward, T.J.; Gorski, L.; Borucki, M.K.; Mandrell, R.E.; Hutchins, J.; Pupedis, K. Intraspecific phylogeny and lineage group identification based on the prfA virulence gene cluster of Listeria monocytogenes. J. Bacteriol. 2004, 186, 4994–5002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haj Hosseini, A.; Sharifan, A.; Tabatabaee, A. Isolation of Listeria monocytogenes from meat and dairy products. J. Med. Microbiol. Infect. Dis. 2014, 2, 159–162. [Google Scholar]
- El-Malek, A.M.A.; Ali, S.F.H.; Hassanein, R.; Mohamed, M.A.; Elsayh, K.I. Occurrence of Listeria species in meat, chicken products and human stools in Assiut city, Egypt with PCR use for rapid identification of Listeria monocytogenes. Vet. World 2010, 3, 353–359. [Google Scholar] [CrossRef]
- Bloom, G.; Merrett, G.B.; Wilkinson, A.; Lin, V.; Paulin, S. Antimicrobial resistance and universal health coverage. BMJ Glob. Health 2017, 2, 1–6. [Google Scholar] [CrossRef]
- Friedman, N.D.; Temkin, E.; Carmeli, Y. The negative impact of antibiotic resistance. Clin. Microbiol. Infect. 2016, 22, 416–422. [Google Scholar] [CrossRef]
- Garedew, L.; Taddese, A.; Biru, T.; Nigatu, S.; Kebede, E.; Ejo, M.; Fikru, A.; Birhanu, T. Prevalence and antimicrobial susceptibility profile of Listeria species from ready-to-eat foods of animal origin in Gondar Town, Ethiopia. BMC Microbiol. 2015, 15, 100. [Google Scholar] [CrossRef] [Green Version]
- Monufia Governorate—Wikipedia. Available online: https://en.wikipedia.org/wiki/Monufia_Governorate (accessed on 21 March 2021).
- Scotter, S.L.; Langton, S.; Lombard, B.; Schulten, S.; Nagelkerke, N.; In’t Veld, P.H.; Rollier, P.; Lahellec, C. Validation of ISO method 11290. Part 1—Detection of Listeria monocytogenes in foods. Int. J. Food Microbiol. 2001, 64, 295–306. [Google Scholar] [CrossRef]
- Wilkinson, H.W. CAMP-disk test for presumptive identification of group B streptococci. J. Clin. Microbiol. 1977, 6, 42–45. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing; CLSI: Tucson, AZ, USA, 2017; ISBN 978-1-56238-804-1. [Google Scholar]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [Green Version]
- Usman, U.B.; Kwaga, J.K.P.; Kabir, J.; Olonitola, O.S.; Radu, S.; Bande, F. Molecular Characterization and Phylogenetic Analysis of Listeria monocytogenes isolated from milk and milk products in Kaduna, Nigeria. Can. J. Infect. Dis. Med. Microbiol. 2016, 2016. [Google Scholar] [CrossRef] [Green Version]
- Hall, T.; Biosciences, I.; Carlsbad, C. BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2011, 2, 60–61. [Google Scholar]
- Ball, N.L.; Adams, C.R.; Xia, W. Overcoming the elusive problem of IS/IT alignment: Conceptual and methodological considerations. In Proceedings of the Ninth Americas Conference on Information Systems, Tampa, FL, USA, 4–6 August 2003; pp. 1669–1676. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Higgins, D.G.; Sharp, P.M. Fast and sensitive multiple sequence alignments on a microcomputer. Bioinformatics 1989, 5, 151–153. [Google Scholar] [CrossRef]
- Kumar, S.; Tamura, K.; Nei, M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 2004, 5, 150–163. [Google Scholar] [CrossRef] [Green Version]
- Taha Said Ahmed, S.S.; Ahmed Tayeb, B. Isolation and molecular detection of Listeria monocytogenes in minced meat, frozen chicken and cheese in Duhok Province, Kurdistan region of Iraq. J. Food Microbiol. Saf. Hyg. 2017, 2, 10–13. [Google Scholar] [CrossRef]
- El-Demerdash, A.S.; Raslan, M.T. Molecular characterization of Listeria monocytogenes isolated from different animal-origin food items from urban and rural areas. Adv. Anim. Vet. Sci. 2019, 7, 51–56. [Google Scholar] [CrossRef] [Green Version]
- Gelbíčová, T.; Karpíšková, R. Occurrence and characteristics of Listeria monocytogenes in ready-to-eat food from retail market in the Czech Republic. Czech. J. Food Sci. 2009, 27, 3–7. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, T.S.; Varjão, L.M.; da Silva, L.N.N.; de Castro Lisboa Pereira, R.; Hofer, E.; Vallim, D.C.; de Castro Almeida, R.C. Listeria monocytogenes at chicken slaughter house: Occurrence, genetic relationship among isolates and evaluation of antimicrobial susceptibility. Food Control. 2018, 88, 131–138. [Google Scholar] [CrossRef]
- Mena, C.; Almeida, G.; Carneiro, L.; Teixeira, P.; Hogg, T.; Gibbs, P.A. Incidence of Listeria monocytogenes in different food products commercialized in Portugal. Food Microbiol. 2004, 21, 213–216. [Google Scholar] [CrossRef]
- Akrami-Mohajeri, F.; Derakhshan, Z.; Ferrante, M.; Hamidiyan, N.; Soleymani, M.; Conti, G.O.; Tafti, R.D. The prevalence and antimicrobial resistance of Listeria spp in raw milk and traditional dairy products delivered in Yazd, Central Iran (2016). Food Chem. Toxicol. 2018, 114, 141–144. [Google Scholar] [CrossRef]
- Basha, K.A.; Kumar, N.R.; Das, V.; Reshmi, K.; Rao, B.M.; Lalitha, K.V.; Joseph, T.C. Prevalence, molecular characterization, genetic heterogeneity and antimicrobial resistance of Listeria monocytogenes associated with fish and fishery environment in Kerala, India. Lett. Appl. Microbiol. 2019, 69, 286–293. [Google Scholar] [CrossRef]
- Girma, Y. Isolation, Identification and antimicrobial susceptibility of Listeria species from raw bovine milk in Debre-Birhan Town, Ethiopia. Ethiop. J. Zoonotic Dis. Public Health 2018, 2, 4. [Google Scholar]
- Somer, L.; Kashi, Y. A PCR method based on 16S rRNA sequence for simultaneous detection of the genus Listeria and the species Listeria monocytogenes in food products. J. Food Prot. 2003, 66, 1658–1665. [Google Scholar] [CrossRef]
- Harb, O.; Elbab, G.; Shawish, R.; Mousa, W.; Abdeen, E. Genetic detection of Listeria monocytogenes recovered from fillet fish samples. Alexandria J. Vet. Sci. 2020, 67, 74. [Google Scholar] [CrossRef]
- Abdellrazeq, G.; Kamar, A.; ElHoushy, S. Molecular characterization of Listeria species isolated from frozen fish. Alexandria J. Vet. Sci. 2014, 40, 1. [Google Scholar] [CrossRef] [Green Version]
- Kaur, S.; Singh, R.; Sran, M.K.; Gill, J.P.S. Molecular characterization of Listeria monocytogenes in white meat samples from Punjab, India. Indian J. Anim. Res. 2018, 52, 1635–1641. [Google Scholar] [CrossRef] [Green Version]
- Soni, D.K.; Dubey, S.K. Phylogenetic analysis of the Listeria monocytogenes based on sequencing of 16S rRNA and hlyA genes. Mol. Biol. Rep. 2014, 41, 8219–8229. [Google Scholar] [CrossRef] [PubMed]
- Soni, D.K.; Singh, M.; Singh, D.V.; Dubey, S.K. Virulence and genotypic characterization of Listeria monocytogenes isolated from vegetable and soil samples. BMC Microbiol. 2014, 14, 241. [Google Scholar] [CrossRef] [Green Version]
- CFSPH. Listeriosis; The Centre for Food Security and Public Health, Iowa State University, College of Veterinary Medicine: Ames, IA, USA, 2019; pp. 1–12. [Google Scholar]





| Targets | Primers | Sequences (5′ to 3′) | Amplicon Size (bp) | Reference |
|---|---|---|---|---|
| 16S rRNA | Forward | CTC CAT AAA GGT GAC CCT | 938 | [18] |
| Reverse | CAG CMG CCG CGG TAA TWC | |||
| hlyA | Forward | CCT AAG ACG CCA ATC GAA | 702 | |
| Reverse | AAG CGCTTG CAA CTG CTC | |||
| inlA | Forward | AGA TCT AGA CCA AGT TAC AAC GCT TCA | 255 | |
| Reverse | TAA TAT CAT TTG CTG TTT TAT CTG TC | |||
| actA | Forward | ACG TGA AGT AAG TCACGT GAT ATT G | 268 | |
| Reverse | ACG TGA AGT AAG CTC ACG TGA TAT TG | |||
| prfA | Forward | ACC GCT CAG AAA AGT TCT TC | 1060 | |
| Reverse | TCT TGT TCT ATT ATGTCT AGC | |||
| iap | Forward | ACA AGC TGC ACC TGT TGC AG | 131 | |
| Reverse | TGA CAG CGT TGT TAG TAG CA |
| Raw Milk (n = 50) | Ice Cream (n = 50) | Minced Meat (n = 50) | Fish Fillet (n = 50) | Sausage (n = 50) | |||||
|---|---|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | No. | % | No. | % |
| 3 | 6 | 0 | 0 | 7 | 14 | 4 | 8 | 3 | 6 |
| Antibiotics | Antimicrobial Classes | Resistant | Intermediate | Sensitive | |||
|---|---|---|---|---|---|---|---|
| No | % | No | % | No | % | ||
| Amoxicillin-Clavulanic acid (AMC) 20/10 µg | β-lactams | 3 | 17.7 | 1 | 5.9 | 13 | 76.4 |
| Cefotaxime (CTX) 30 µg | β-lactams | 3 | 17.7 | 2 | 11.8 | 12 | 70.5 |
| Amoxicillin (AX) 30 µg | β-lactams | 5 | 29.5 | 1 | 5.9 | 11 | 64.6 |
| Norfloxacin (NOR) 10 µg | Fluoroquinolones | 2 | 11.8 | 1 | 5.9 | 14 | 82.3 |
| Ciprofloxacin (CIP) 5 µg | Fluoroquinolones | 5 | 29.4 | 2 | 11.8 | 10 | 58.8 |
| Levofloxacin (LEV) 5 µg | Fluoroquinolones | 7 | 41.2 | 2 | 11.8 | 8 | 47 |
| Danofloxacin (DA) 2 µg | Fluoroquinolones | 7 | 41.2 | 2 | 11.8 | 8 | 47 |
| Nalidixic acid (NA) 30 µg | Fluoroquinolones | 3 | 17.7 | 10 | 58.8 | 4 | 23.5 |
| Amikacin (AK) 30 µg | Aminoglycosides | 5 | 29.4 | 1 | 5.9 | 11 | 64.6 |
| Gentamicin (CN) 10 µg | Aminoglycosides | 5 | 29.5 | 2 | 11.8 | 10 | 58.7 |
| Erythromycin (E) 15 µg | Macrolides | 5 | 29.5 | 1 | 5.9 | 11 | 64.6 |
| Azithromycin (AZM) 15 µg | Macrolides | 7 | 41.2 | 7 | 41.2 | 3 | 17.7 |
| Doxycycline (DO) 30 µg | Tetracycline | 11 | 64.6 | 3 | 17.7 | 3 | 17.7 |
| Oxytetracycline (T) 30 µg | Tetracycline | 13 | 76.4 | 1 | 5.9 | 3 | 17.7 |
| Chloramphenicol (C) 30 µg | Chloramphenicol | 12 | 70.5 | 2 | 11.8 | 3 | 17.7 |
| Trimethoprim-Sulfamethoxazole (SXT) 12.5/23.75 µg | Sulfonamides | 13 | 76.4 | 2 | 11.8 | 2 | 11.8 |
| Vancomycin (VA) 30 µg | Glycopeptides | 5 | 29.5 | 2 | 11.8 | 10 | 58.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdeen, E.E.; Mousa, W.S.; Harb, O.H.; Fath-Elbab, G.A.; Nooruzzaman, M.; Gaber, A.; Alsanie, W.F.; Abdeen, A. Prevalence, Antibiogram and Genetic Characterization of Listeria monocytogenes from Food Products in Egypt. Foods 2021, 10, 1381. https://0-doi-org.brum.beds.ac.uk/10.3390/foods10061381
Abdeen EE, Mousa WS, Harb OH, Fath-Elbab GA, Nooruzzaman M, Gaber A, Alsanie WF, Abdeen A. Prevalence, Antibiogram and Genetic Characterization of Listeria monocytogenes from Food Products in Egypt. Foods. 2021; 10(6):1381. https://0-doi-org.brum.beds.ac.uk/10.3390/foods10061381
Chicago/Turabian StyleAbdeen, Eman E., Walid S. Mousa, Ola. H. Harb, Gehad A. Fath-Elbab, Mohammed Nooruzzaman, Ahmed Gaber, Walaa F. Alsanie, and Ahmed Abdeen. 2021. "Prevalence, Antibiogram and Genetic Characterization of Listeria monocytogenes from Food Products in Egypt" Foods 10, no. 6: 1381. https://0-doi-org.brum.beds.ac.uk/10.3390/foods10061381