Modulation of Designed Gut Bacterial Communities by Prebiotics and the Impact of Their Metabolites on Intestinal Cells
Abstract
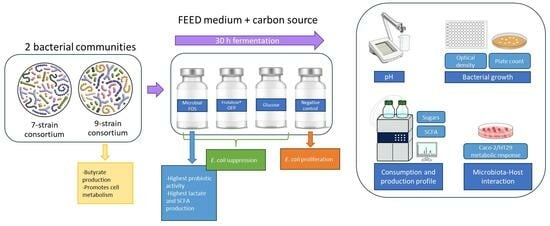
:1. Introduction
2. Materials and Methods
2.1. Carbohydrates Tested
2.2. Microbial Community
2.3. Fermentation Assays
2.3.1. Bacterial Growth Analysis
2.3.2. Carbohydrates and Organic Acids Analysis
2.4. Cell Culture Experiments
Cell Metabolic Activity Assay
2.5. Statistical Analysis
3. Results
3.1. Bacterial Growth and pH Profile
3.2. Carbohydrate Consumption
3.3. Short-Chain Fatty Acid Production
3.4. Effect of Metabolites Produced by Different Microbial Communities Using Distinct Carbon Sources on an Intestinal Cell Co-Culture
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What Is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Stojanov, S.; Berlec, A.; Štrukelj, B. The Influence of Probiotics on the Firmicutes/Bacteroidetes Ratio in the Treatment of Obesity and Inflammatory Bowel Disease. Microorganisms 2020, 8, 1715. [Google Scholar] [CrossRef] [PubMed]
- Arboleya, S.; Watkins, C.; Stanton, C.; Ross, R.P. Gut Bifidobacteria Populations in Human Health and Aging. Front. Microbiol. 2016, 7, 1204. [Google Scholar] [CrossRef] [PubMed]
- Flint, H.J.; Jude, N. Role of Microbes in Carbohydrate Digestion. Food Sci. Technol. 2015, 29, 24–26. [Google Scholar]
- Jovel, J.; Dieleman, L.A.; Kao, D.; Mason, A.L.; Wine, E. The Human Gut Microbiome in Health and Disease. In Metagenomics: Perspectives, Methods, and Applications; Academic Press: Cambridge, MA, USA, 2018; Volume 13, pp. 197–213. ISBN 9780128134030. [Google Scholar]
- Slavin, J. Fiber and Prebiotics: Mechanisms and Health Benefits. Nutrients 2013, 5, 1417–1435. [Google Scholar] [CrossRef]
- Hiippala, K.; Jouhten, H.; Ronkainen, A.; Hartikainen, A.; Kainulainen, V.; Jalanka, J.; Satokari, R. The Potential of Gut Commensals in Reinforcing Intestinal Barrier Function and Alleviating Inflammation. Nutrients 2018, 10, 988. [Google Scholar] [CrossRef]
- Ghosh, S.S.; Wang, J.; Yannie, P.J.; Ghosh, S. Intestinal Barrier Dysfunction, LPS Translocation, and Disease Development. J. Endocr. Soc. 2020, 4, bvz039. [Google Scholar] [CrossRef]
- Ducatelle, R.; Eeckhaut, V.; Haesebrouck, F.; Van Immerseel, F. A Review on Prebiotics and Probiotics for the Control of Dysbiosis: Present Status and Future Perspectives. Animal 2014, 9, 43–48. [Google Scholar] [CrossRef]
- Collins, S.M. A Role for the Gut Microbiota in IBS. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 497–505. [Google Scholar] [CrossRef]
- Everard, A.; Cani, P.D. Diabetes, Obesity and Gut Microbiota. Best. Pract. Res. Clin. Gastroenterol. 2013, 27, 73–83. [Google Scholar] [CrossRef]
- Joossens, M.; Huys, G.; Cnockaert, M.; De Preter, V.; Verbeke, K.; Rutgeerts, P.; Vandamme, P.; Vermeire, S. Dysbiosis of the Faecal Microbiota in Patients with Crohn’s Disease and Their Unaffected Relatives. Gut 2011, 60, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Petersen, E.B.M.; Skov, L.; Thyssen, J.P.; Jensen, P. Role of the Gut Microbiota in Atopic Dermatitis: A Systematic Review. Acta Derm. Venereol. 2018, 99, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. Expert Consensus Document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) Consensus Statement on the Definition and Scope of Prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Nobre, C.; Simões, L.S.; Gonçalves, D.A.; Berni, P.; Teixeira, J.A. Fructooligosaccharides Production and the Health Benefits of Prebiotics. In Current Developments in Biotechnology and Bioengineering: Technologies for Production of Nutraceuticals and Functional Food Products; Elsevier: Amsterdam, The Netherlands, 2022; pp. 109–138. [Google Scholar] [CrossRef]
- Scheid, M.M.A.; Moreno, Y.M.F.; Maróstica Junior, M.R.; Pastore, G.M. Effect of Prebiotics on the Health of the Elderly. Food Res. Int. 2013, 53, 426–432. [Google Scholar] [CrossRef]
- Rose, E.C.; Odle, J.; Blikslager, A.T.; Ziegler, A.L. Probiotics, Prebiotics and Epithelial Tight Junctions: A Promising Approach to Modulate Intestinal Barrier Function. Int. J. Mol. Sci. 2021, 22, 6729. [Google Scholar] [CrossRef]
- Knudsen, K.E.B.; Lærke, H.N.; Hedemann, M.S.; Nielsen, T.S.; Ingerslev, A.K.; Nielsen, D.S.G.; Theil, P.K.; Purup, S.; Hald, S.; Schioldan, A.G.; et al. Impact of Diet-Modulated Butyrate Production on Intestinal Barrier Function and Inflammation. Nutrients 2018, 10, 1499. [Google Scholar] [CrossRef]
- Van der Beek, C.M.; Dejong, C.H.C.; Troost, F.J.; Masclee, A.A.M.; Lenaerts, K. Role of Short-Chain Fatty Acids in Colonic Inflammation, Carcinogenesis, and Mucosal Protection and Healing. Nutr. Rev. 2017, 75, 286–305. [Google Scholar] [CrossRef]
- Portincasa, P.; Bonfrate, L.; Vacca, M.; De Angelis, M.; Farella, I.; Lanza, E.; Khalil, M.; Wang, D.Q.H.; Sperandio, M.; Di Ciaula, A. Gut Microbiota and Short Chain Fatty Acids: Implications in Glucose Homeostasis. Int. J. Mol. Sci. 2022, 23, 1105. [Google Scholar] [CrossRef]
- Wu, Y.; Xu, H.; Tu, X.; Gao, Z. The Role of Short-Chain Fatty Acids of Gut Microbiota Origin in Hypertension. Front. Microbiol. 2021, 12, 730809. [Google Scholar] [CrossRef]
- Rios-Covian, D.; Salazar, N.; Gueimonde, M.; de los Reyes-Gavilan, C.G. Shaping the Metabolism of Intestinal Bacteroides Population through Diet to Improve Human Health. Front. Microbiol. 2017, 8, 376. [Google Scholar] [CrossRef]
- Sarbini, S.; Rastall, R. Prebiotics: Metabolism, Structure, and Function. Funct. Food Rev. 2011, 3, 93–106. [Google Scholar] [CrossRef]
- Pham, V.T.; Seifert, N.; Richard, N.; Raederstorff, D.; Steinert, R.; Prudence, K.; Hasan Mohajeri, M. The Effects of Fermentation Products of Prebiotic Fibres on Gut Barrier and Immune Functions. Vitro PeerJ 2018, 2018, e5288. [Google Scholar] [CrossRef] [PubMed]
- Nobre, C.; Gonçalves, D.A.; Teixeira, J.A.; Rodrigues, L.R. One-Step Co-Culture Fermentation Strategy to Produce High-Content Fructo-Oligosaccharides. Carbohydr. Polym. 2018, 201, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Roupar, D.; Coelho, M.C.; Gonçalves, D.A.; Silva, S.P.; Coelho, E.; Silva, S.; Coimbra, M.A.; Pintado, M.; Teixeira, J.A.; Nobre, C. Evaluation of Microbial-Fructo-Oligosaccharides Metabolism by Human Gut Microbiota Fermentation as Compared to Commercial Inulin-Derived Oligosaccharides. Foods 2022, 11, 954. [Google Scholar] [CrossRef] [PubMed]
- Sivieri, K.; Morales, M.L.V.; Adorno, M.A.T.; Sakamoto, I.K.; Saad, S.M.I.; Rossi, E.A. Lactobacillus acidophilus CRL 1014 Improved “Gut Health” in the SHIME® Reactor. BMC Gastroenterol. 2013, 13, 100. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, G.T.; Hay, S.; Macfarlane, S.; Gibson, G.R. Effect of Different Carbohydrates on Growth, Polysaccharidase and Glycosidase Production by Bacteroides ovatus, in Batch and Continuous Culture. J. Appl. Bacteriol. 1990, 68, 179–187. [Google Scholar] [CrossRef]
- Allison, C.; McFarlan, C.; Macfarlane, G.T. Studies on Mixed Populations of Human Intestinal Bacteria Grown in Single-Stage and Multistage Continuous Culture Systems. Appl. Environ. Microbiol. 1989, 55, 672–678. [Google Scholar] [CrossRef]
- Beerens, H. Detection of Bifidobacteria by Using Propionic Acid as a Selective Agent. Appl. Environ. Microbiol. 1991, 57, 2418–2419. [Google Scholar] [CrossRef]
- O’Brien, J.; Wilson, I.; Orton, T.; Pognan, F. Investigation of the Alamar Blue (Resazurin) Fluorescent Dye for the Assessment of Mammalian Cell Cytotoxicity. Eur. J. Biochem. 2000, 267, 5421–5426. [Google Scholar] [CrossRef]
- Morrison, D.J.; Preston, T. Formation of Short Chain Fatty Acids by the Gut Microbiota and Their Impact on Human Metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef]
- Venegas, D.P.; De La Fuente, M.K.; Landskron, G.; González, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs) Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Kwon, O.; Ryu, T.Y.; Jung, C.R.; Kim, J.; Min, J.K.; Kim, D.S.; Son, M.Y.; Cho, H.S. Propionate of a Microbiota Metabolite Induces Cell Apoptosis and Cell Cycle Arrest in Lung Cancer. Mol. Med. Rep. 2019, 20, 1569–1574. [Google Scholar] [CrossRef]
- Tian, Y.; Xu, Q.; Sun, L.; Ye, Y.; Ji, G. Short-Chain Fatty Acids Administration Is Protective in Colitis-Associated Colorectal Cancer Development. J. Nutr. Biochem. 2018, 57, 103–109. [Google Scholar] [CrossRef]
- Wu, X.; Wu, Y.; He, L.; Wu, L.; Wang, X.; Liu, Z. Effects of the Intestinal Microbial Metabolite Butyrate on the Development of Colorectal Cancer. J. Cancer 2018, 9, 2510–2517. [Google Scholar] [CrossRef] [PubMed]
- Ashaolu, T.J.; Ashaolu, J.O.; Adeyeye, S.A.O. Fermentation of Prebiotics by Human Colonic Microbiota in Vitro and Short-Chain Fatty Acids Production: A Critical Review. J. Appl. Microbiol. 2021, 130, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Kettle, H.; Donnelly, R.; Flint, H.J.; Marion, G. PH Feedback and Phenotypic Diversity within Bacterial Functional Groups of the Human Gut. J. Theor. Biol. 2014, 342, 62–69. [Google Scholar] [CrossRef]
- Nobre, C.; Sousa, S.C.; Silva, S.P.; Pinheiro, A.C.; Coelho, E.; Vicente, A.A.; Gomes, A.M.P.; Coimbra, M.A.; Teixeira, J.A.; Rodrigues, L.R. In Vitro Digestibility and Fermentability of Fructo-Oligosaccharides Produced by Aspergillus ibericus. J. Funct. Foods 2018, 46, 278–287. [Google Scholar] [CrossRef]
- Ito, H.; Takemura, N.; Sonoyama, K.; Kawagishi, H.; Topping, D.L.; Conlon, M.A.; Morita, T. Degree of Polymerization of Inulin-Type Fructans Differentially Affects Number of Lactic Acid Bacteria, Intestinal Immune Functions, and Immunoglobulin a Secretion in the Rat Cecum. J. Agric. Food Chem. 2011, 59, 5771–5778. [Google Scholar] [CrossRef]
- Endo, H.; Tamura, K.; Fukasawa, T.; Kanegae, M.; Koga, J. Comparison of Fructooligosaccharide Utilization by Lactobacillus and Bacteroides Species. Biosci. Biotechnol. Biochem. 2012, 76, 176–179. [Google Scholar] [CrossRef]
- Wong, C.B.; Odamaki, T.; Xiao, J.Z. Insights into the Reason of Human-Residential Bifidobacteria (HRB) Being the Natural Inhabitants of the Human Gut and Their Potential Health-Promoting Benefits. FEMS Microbiol. Rev. 2020, 44, 369–385. [Google Scholar] [CrossRef]
- Reichardt, N.; Vollmer, M.; Holtrop, G.; Farquharson, F.M.; Wefers, D.; Bunzel, M.; Duncan, H.S.; Drew, E.J.; Williams, M.L.; Milligan, G.; et al. Specific substrate-driven changes in human faecal microbiota composition contrast with functional redundancy in short-chain fatty acid production. ISME J. 2018, 12, 610–622. [Google Scholar] [CrossRef] [PubMed]
- Duncan, S.H.; Louis, P.; Thomson, J.M.; Flint, H.J. The Role of PH in Determining the Species Composition of the Human Colonic Microbiota. Environ. Microbiol. 2009, 11, 2112–2122. [Google Scholar] [CrossRef] [PubMed]
- Barroso, E.; Montilla, A.; Corzo, N.; Peláez, C.; Martínez-Cuesta, M.C.; Requena, T. Effect of Lactulose-Derived Oligosaccharides on Intestinal Microbiota during the Shift between Media with Different Energy Contents. Food Res. Int. 2016, 89, 302–308. [Google Scholar] [CrossRef]
- González, A.; Conceição, E.; Teixeira, J.A.; Nobre, C. In Vitro Models as a Tool to Study the Role of Gut Microbiota in Obesity. Crit. Rev. Food Sci. Nutr. 2023, 66, 2682–2684. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, H.; Hutkins, R.W. Fermentation of Fructooligosaccharides by Lactic Acid Bacteria and Bifidobacteria. Appl. Environ. Microbiol. 2000, 66, 2682–2684. [Google Scholar] [CrossRef]
- Sonnenburg, E.D.; Zheng, H.; Joglekar, P.; Higginbottom, S.K.; Firbank, S.J.; Bolam, D.N.; Sonnenburg, J.L. Specificity of Polysaccharide Use in Intestinal Bacteroides Species Determines Diet-Induced Microbiota Alterations. Cell 2010, 141, 1241–1252. [Google Scholar] [CrossRef]
- Der Van Meulen, R.; Makras, L.; Verbrugghe, K.; Adriany, T.; De Vuyst, L. In Vitro Kinetic Analysis of Oligofructose Consumption by Bacteroides and Bifidobacterium spp. Indicates Different Degradation Mechanisms. Appl. Environ. Microbiol. 2006, 72, 1006–1012. [Google Scholar] [CrossRef]
- Sannohe, Y.; Fukasawa, T.; Koga, J.; Kubota, H.; Kanegae, M. Comparison of the Growth of Bifidobacteria in Two Culture Media Containing Either 1-Kestose (GF2) or Nystose (GF3). Biosci. Microflora 2008, 27, 13–17. [Google Scholar] [CrossRef]
- Suzuki, N.; Aiba, Y.; Takeda, H.; Fukumori, Y.; Koga, Y. Superiority of 1-Kestose, the Smallest Fructo-Oligosaccharide, to a Synthetic Mixture of Fructo-Oligosaccharides in the Selective Stimulating Activity on Bifidobacteria. Biosci. Microflora 2006, 25, 109–116. [Google Scholar] [CrossRef]
- Biedrzycka, E.; Bielecka, M. Prebiotic Effectiveness of Fructans of Different Degrees of Polymerization. Trends Food Sci. Technol. 2004, 15, 170–175. [Google Scholar] [CrossRef]
- Rossi, M.; Corradini, C.; Amaretti, A.; Nicolini, M.; Pompei, A.; Zanoni, S.; Matteuzzi, D. Fermentation of Fructooligosaccharides and Inulin by Bifidobacteria: A Comparative Study of Pure and Fecal Cultures. Appl. Environ. Microbiol. 2005, 71, 6150–6158. [Google Scholar] [CrossRef] [PubMed]
- Hernot, D.C.; Boileau, T.W.; Bauer, L.L.; Middelbos, I.S.; Murphy, M.R.; Swanson, K.S.; Fahey, G.C. In Vitro Fermentation Profiles, Gas Production Rates, and Microbiota Modulation as Affected by Certain Fructans, Galactooligosaccharides, and Polydextrose. J. Agric. Food Chem. 2009, 57, 1354–1361. [Google Scholar] [CrossRef] [PubMed]
- Bajury, D.M.; Nashri, S.M.; King Jie Hung, P.; Sarbini, S.R. Evaluation of Potential Prebiotics: A Review. Food Rev. Int. 2018, 34, 639–664. [Google Scholar] [CrossRef]
- Pacifici, R.; Lawenius, L.; Sjögren, K.; Ohlsson, C. Bone and the Microbiome. In Marcus and Feldman’s Osteoporosis; Elsevier: Amsterdam, The Netherlands, 2020; pp. 969–988. ISBN 9780128130735. [Google Scholar]
- Louis, P.; Flint, H.J. Formation of Propionate and Butyrate by the Human Colonic Microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Iraporda, C.; Errea, A.; Romanin, D.E.; Cayet, D.; Pereyra, E.; Pignataro, O.; Sirard, J.C.; Garrote, G.L.; Abraham, A.G.; Rumbo, M. Lactate and Short Chain Fatty Acids Produced by Microbial Fermentation Downregulate Proinflammatory Responses in Intestinal Epithelial Cells and Myeloid Cells. Immunobiology 2015, 220, 1161–1169. [Google Scholar] [CrossRef] [PubMed]
- Matsuki, T.; Pédron, T.; Regnault, B.; Mulet, C.; Hara, T.; Sansonetti, P.J. Epithelial Cell Proliferation Arrest Induced by Lactate and Acetate from Lactobacillus casei and Bifidobacterium breve. PLoS ONE 2013, 8, e63053. [Google Scholar] [CrossRef]
- Lee, Y.S.; Kim, T.Y.; Kim, Y.; Lee, S.H.; Kim, S.; Kang, S.W.; Yang, J.Y.; Baek, I.J.; Sung, Y.H.; Park, Y.Y.; et al. Microbiota-Derived Lactate Accelerates Intestinal Stem-Cell-Mediated Epithelial Development. Cell Host Microbe 2018, 24, 833–846.e6. [Google Scholar] [CrossRef]
- Parente, I.A.; Xavier, M.; Roupar, D.; Amado, I.R.; Berni, P.; Botelho, C.; Teixeira, J.A.; Pastrana, L.; Nobre, C.; Gonçalves, C. Effect of Prebiotic Fermentation Products from Primary Human Gut Microbiota on an In Vitro Intestinal Model. J. Funct. Foods 2022, 96, 105200. [Google Scholar] [CrossRef]
- Verbeke, K.A.; Boobis, A.R.; Chiodini, A.; Edwards, C.A.; Franck, A.; Kleerebezem, M.; Nauta, A.; Raes, J.; Van Tol, E.A.F.; Tuohy, K.M. Towards Microbial Fermentation Metabolites as Markers for Health Benefits of Prebiotics. Nutr. Res. Rev. 2015, 28, 42–66. [Google Scholar] [CrossRef]
- Hamer, H.M.; Jonkers, D.; Venema, K.; Vanhoutvin, S.; Troost, F.J.; Brummer, R.J. Review Article: The Role of Butyrate on Colonic Function. Aliment. Pharmacol. Ther. 2008, 27, 104–119. [Google Scholar] [CrossRef]






| Bacterial Strain | Culture Media * |
|---|---|
| Bacteroides dorei DSM 17855 | BHIS |
| Bacteroides vulgatus DSM 1447 | |
| Bacteroides thetaiotaomicron DSM 2079 | |
| Bifidobacterium adolescentis CECT 5781 | BSM |
| Bifidobacterium longum DSM 20219 | |
| Lactobacillus acidophilus CECT 288 | MRS |
| Lactobacillus rhamnosus ATCC 4356 | |
| Roseburia faecis DSM 16840 | RBM:330 |
| Escherichia coli CECT 736 | NB |
| SCFA (g·L−1) | Time (h) | Glucose | Microbial-FOS | Frutalose® OFP | Negative Control 1 | ||||
|---|---|---|---|---|---|---|---|---|---|
| 7SC | 9SC | 7SC | 9SC | 7SC | 9SC | 7SC | 9SC | ||
| Acetate | 0 | 0.31 ± 0.00 a | 0.49 ± 0.01 b* | 0.31 ± 0.00 a | 0.49 ± 0.01 b* | 0.31 ± 0.00 a | 0.49 ± 0.01 b* | 0.31 ± 0.00 a | 0.49 ± 0.01 b* |
| 12 | 0.81 ± 0.03 b | 0.83 ± 0.02 a | 0.66 ± 0.03 a | 0.97 ± 0.04 b | 0.72 ± 0.01 a | 2.30 ± 0.02 d* | 0.98 ± 0.02 c | 1.63 ± 0.05 c* | |
| 30 | 0.84 ± 0.01 a | 1.04 ± 0.05 a | 1.20 ± 0.01 b | 1.01 ± 0.10 a | 0.81 ± 0.01 a | 2.59 ± 0.08 c* | 0.89 ± 0.04 a | 1.36 ± 0.04 b* | |
| Propionate | 0 | 1.98 ± 0.06 a | 2.68 ± 0.07 b* | 1.98 ± 0.06 a | 2.68 ± 0.07 b* | 1.98 ± 0.06 a | 2.68 ± 0.07 b* | 1.98 ± 0.06 a | 2.68 ± 0.07 b* |
| 12 | 2.93 ± 0.04 c | 3.82 ± 0.05 c* | 2.66 ± 0.06 b | 3.92 ± 0.13 c* | 2.44 ± 0.02 a | 3.31 ± 0.06 b* | 2.65 ± 0.03 b | 2.81 ± 0.04 a* | |
| 30 | 3.05 ± 0.05 d | 3.81 ± 0.14 b* | 2.77 ± 0.05 c | 3.85 ± 0.11 b* | 2.48 ± 0.00 b | 2.84 ± 0.08 a* | 1.96 ± 0.03 a | 3.15 ± 0.07 a* | |
| Butyrate | 0 | 0.12 ± 0.00 a | 1.17 ± 0.02 b* | 0.12 ± 0.00 a | 1.17 ± 0.02 b* | 0.12 ± 0.00 a | 1.17 ± 0.02 b* | 0.12 ± 0.00 a | 1.17 ± 0.02 b* |
| 12 | 0.18 ± 0.00 c | 1.58 ± 0.14 a* | 0.13 ± 0.01 b | 2.71 ± 0.07 c* | 0.18 ± 0.00 c | 1.88 ± 0.03 b* | 0.07 ± 0.00 a | 1.33 ± 0.05 a* | |
| 30 | 0.18 ± 0.00 c | 1.66 ± 0.08 a* | 0.13 ± 0.00 b | 2.97 ± 0.06 c* | 0.18 ± 0.00 c | 2.26 ± 0.03 b* | 0.07 ± 0.00 a | 1.52 ± 0.06 a* | |
| Lactate | 0 | 4.43 ± 0.07 a | 4.71 ± 0.13 a | 4.43 ± 0.07 a | 4.71 ± 0.13 a | 4.43 ± 0.07 a | 4.71 ± 0.13 a | 4.43 ± 0.07 a | 4.71 ± 0.13 a |
| 12 | 7.71 ± 0.04 d | 10.59 ± 0.15 c* | 6.72 ± 0.07 c | 8.94 ± 0.23 b* | 4.53 ± 0.04 b | 14.00 ± 0.18 d* | 3.60 ± 0.04 a | 5.20 ± 0.05 a* | |
| 30 | 9.87 ± 0.08 c | 15.11 ± 0.13 d* | 12.17 ± 0.01 d | 10.88 ± 0.20 b* | 5.93 ± 0.01 b | 14.07 ± 0.11 c* | 3.73 ± 0.04 a | 8.47 ± 0.16 a* | |
| Total SCFA | 0 | 2.41 ± 0.06 a | 4.34 ± 0.09 b* | 2.41 ± 0.06 a | 4.34 ± 0.09 b* | 2.41 ± 0.06 a | 4.34 ± 0.09 b* | 2.41 ± 0.06 a | 4.34 ± 0.09 b* |
| 12 | 3.91 ± 0.07 c | 6.23 ± 0.14 a* | 3.45 ± 0.08 a | 7.60 ± 0.23 b* | 3.34 ± 0.02 a | 7.48 ± 0.07 b* | 3.70 ± 0.05 b | 5.77 ± 0.14 a* | |
| 30 | 4.08 ± 0.06 c | 6.51 ± 0.23 a* | 4.11 ± 0.04 c | 7.83 ± 0.20 b* | 3.48 ± 0.01 b | 7.70 ± 0.13 b* | 2.92 ± 0.03 a | 6.03 ± 0.13 a* | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roupar, D.; González, A.; Martins, J.T.; Gonçalves, D.A.; Teixeira, J.A.; Botelho, C.; Nobre, C. Modulation of Designed Gut Bacterial Communities by Prebiotics and the Impact of Their Metabolites on Intestinal Cells. Foods 2023, 12, 4216. https://0-doi-org.brum.beds.ac.uk/10.3390/foods12234216
Roupar D, González A, Martins JT, Gonçalves DA, Teixeira JA, Botelho C, Nobre C. Modulation of Designed Gut Bacterial Communities by Prebiotics and the Impact of Their Metabolites on Intestinal Cells. Foods. 2023; 12(23):4216. https://0-doi-org.brum.beds.ac.uk/10.3390/foods12234216
Chicago/Turabian StyleRoupar, Dalila, Abigail González, Joana T. Martins, Daniela A. Gonçalves, José A. Teixeira, Cláudia Botelho, and Clarisse Nobre. 2023. "Modulation of Designed Gut Bacterial Communities by Prebiotics and the Impact of Their Metabolites on Intestinal Cells" Foods 12, no. 23: 4216. https://0-doi-org.brum.beds.ac.uk/10.3390/foods12234216