Isolation and Characterization of Cryotolerant Yeasts from Fiano di Avellino Grapes Fermented at Low Temperatures
Abstract
:1. Introduction
2. Materials and Methods
2.1. Origin of Grape Samples and Fermentation Conditions
2.2. Fermentation Monitoring and Yeast Isolation
2.3. Yeast Molecular Identification and Typing
2.4. Technological Characterization of Saccharomyces spp. Strains
2.5. Microfermentation Trials
2.6. VOCs Analysis
2.7. Statistical Analysis
3. Results
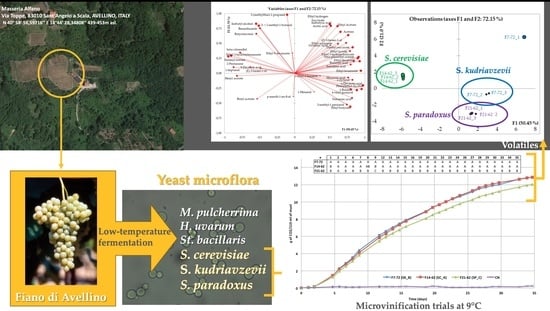
3.1. Fermentation Monitoring and Yeast Isolation
3.2. Yeast Molecular Identification and Typing of Isolates
3.3. Technological Characterization of Saccharomyces spp. Yeast Strains
3.4. Microfermentation Trials
3.5. VOCs Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Molina, A.M.; Swiegers, J.H.; Varela, C.; Pretorius, I.S.; Agosin, E. Influence of wine fermentation temperature on the synthesis of yeast-derived volatile aroma compounds. Appl. Microbiol. Biotechnol. 2007, 77, 675–687. [Google Scholar] [CrossRef]
- Mouret, J.R.; Camarasa, C.; Angenieux, M.; Aguera, E.; Perez, M.; Farines, V.; Sablayrolles, J.M. Kinetic analysis and gas-liquid balances of the production of fermentative aromas during winemaking fermentations: Effect of assimilable nitrogen and temperature. Food Res. Int. 2014, 62, 1–10. [Google Scholar] [CrossRef]
- Masneuf, I.; Hansen, J.; Groth, C.; Piskur, J.; Dubourdieu, D. New hybrids between Saccharomyces sensu stricto yeast species found among wine and cider production strains. Appl. Env. Microbiol. 1998, 64, 3887–3892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez, S.S.; Barrio, E.; Gafner, J.; Querol, A. Natural hybrids from Saccharomyces cerevisiae, Saccharomyces bayanus and Saccharomyces kudriavzevii in wine fermentations. FEMS Yeast Res. 2006, 6, 1221–1234. [Google Scholar] [CrossRef] [Green Version]
- García-Ríos, E.; Guillén, A.; de la Cerda, R.; Pérez-Través, L.; Querol, A.; Guillamón, J.M. Improving the cryotolerance of wine yeast by interspecific hybridization in the genus Saccharomyces. Front. Microbiol. 2019, 9, 3232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aponte, M.; Blaiotta, G. Potential role of yeast strains isolated from grapes in the production of Taurasi DOCG. Front. Microbiol. 2016, 7, 809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Börlin, M.; Miot-Sertier, C.; Vinsonneau, E.; Becquet, S.; Salin, F.; Bely, M.; Lucas, P.; Albertin, W.; Legras, J.-L.; Masneuf-Pomarède, I. The “pied de cuve” as an alternative way to manage indigenous fermentation: Impact on the fermentative process and Saccharomyces cerevisiae diversity. OENO ONE 2020, 3, 435–442. [Google Scholar] [CrossRef]
- Legras, J.L.; Karst, F. Optimisation of interdelta analysis for Saccharomyces cerevisiae strain characterization. FEMS Microbiol. Lett. 2003, 221, 249–255. [Google Scholar] [CrossRef] [Green Version]
- Marinangeli, P.; Angelozzi, D.; Ciani, M.; Clementi, F.; Mannazzu, I. Minisatellites in Saccharomyces cerevisiae genes encoding cell wall proteins: A new way towards wine strain characterization. FEMS Yeast Res. 2004, 4, 427–435. [Google Scholar] [CrossRef] [Green Version]
- Aponte, M.; Blaiotta, G. Selection of an autochthonous Saccharomyces cerevisiae strain for the vinification of “Moscato di Saracena”, a southern Italy (Calabria Region) passito wine. Food Microbiol. 2016, 54, 30–39. [Google Scholar] [CrossRef]
- Fia, G.; Giovani, G.; Rosi, I. Study of beta-glucosidase production by wine-related yeasts during alcoholic fermentation. A new rapid fluorimetric method to determine enzymatic activity. J. Appl. Microbiol. 2005, 99, 509–517. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, L.F.; Espinosa, J.C.; Fernandez-Gonzales, M.; Briones, A. Beta-glucosidase activity in a Saccharomyces cerevisiae wine strain. Int. J. Food Microbiol. 2002, 80, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Slifkin, M. Tween 80 opacity test responses of various Candida species. J. Clin. Microbiol. 2000, 38, 4626–4628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fadda, M.E.; Mossa, V.; Pisano, M.B.; Depilano, M.; Casentino, S. Occurrence and characterization of yeasts isolated from artisanal Fiore Sardo cheese. Int. J. Food Microbiol. 2004, 5, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Romano, P.; Siesto, G.; Capece, A.; Pietrafesa, R.; Lanciotti, R.; Patrignani, F.; Tufariello, M. Validation of a standard protocol to assess the fermentative and chemical properties of Saccharomyces cerevisiae wine strains. Front. Microbiol. 2022, 13, 830277. [Google Scholar] [CrossRef] [PubMed]
- De Filippis, F.; Aponte, M.; Piombino, P.; Lisanti, M.T.; Moio, L.; Ercolinia, D.; Blaiotta, G. Influence of microbial communities on the chemical and sensory features of Falanghina sweet passito wines. Food Res Intern. 2019, 120, 740–747. [Google Scholar] [CrossRef] [PubMed]
- Piombino, P.; Genovese, A.; Gambuti, A.; Lamorte, S.A.; Lisanti, M.T.; Moio, L. Effects of off-vine bunches shading and cryomaceration on free and glycosilated flavours of Malvasia delle Lipari wine. Int. J. Food Sci Technol. 2010, 45, 234–244. [Google Scholar] [CrossRef]
- Mills, D.A.; Phister, T.; Neeley, E.; Johannsen, E. Wine Fermentation. In Molecular Techniques in the Microbial Ecology of Fermented Foods; Cocolin, L., Ercolini, D., Eds.; Springer: New York, NY, USA, 2008; pp. 161–192. [Google Scholar]
- Zilelidou, E.A.; Nisiotou, A. Understanding wine through yeast interactions. Microorganisms 2021, 9, 1620. [Google Scholar] [CrossRef]
- Pérez-Torrado, R.; Barrio, E.; Querol, A. Alternative yeasts for winemaking: Saccharomyces non-cerevisiae and its hybrids. Crit. Rev. Food Sci. Nutr. 2018, 58, 1780–1790. [Google Scholar] [CrossRef] [Green Version]
- Johnson, L.J.; Koufopanou, V.; Goddard, M.R.; Hetherington, R.; Schäfer, S.M.; Burt, A. Population genetics of the wild yeast Saccharomyces paradoxus. Genetics 2004, 166, 43–52. [Google Scholar] [CrossRef]
- Sniegowski, P.D.; Dombrowski, P.G.; Fingerman, E. Saccharomyces cerevisiae and Saccharomyces paradoxus coexist in a natural woodland site in North America and display different levels of reproductive isolation from European conspecifics. FEMS Yeast Res. 2002, 1, 299–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Redzepović, S.; Orlić, S.; Sikora, S.; Majdak, A.; Pretorius, I.S. Identification and characterization of Saccharomyces cerevisiae and Saccharomyces paradoxus strains isolated from Croatian vineyards. Lett. Appl. Microbiol. 2002, 35, 305–310. [Google Scholar] [CrossRef]
- Vaudano, E.; Quinterno, G.; Costantini, A.; Pulcini, L.; Pessione, E.; Garcia-Moruno, E. Yeast distribution in Grignolino grapes growing in a new vineyard in Piedmont and the technological characterization of indigenous Saccharomyces spp. strains. Int. J. Food Microbiol. 2019, 289, 154–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naumov, G.I.; James, S.A.; Naumova, E.S.; Louis, E.J.; Roberts, I.N. Three new species in the Saccharomyces sensu stricto complex: Saccharomyces cariocanus, Saccharomyces kudriavzevii and Saccharomyces mikatae. Int. J. Sys. Evol. Microbiol. 2000, 39050, 1931–1942. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, J.; Richards, K.; Niederer, H.; Lee, S.A.; Dunbar, P.R.; Gardner, R.C. A homozygous diploid subset of commercial wine yeast strains. Antonie Leeuwenhoek 2006, 89, 27–37. [Google Scholar] [CrossRef] [PubMed]
- González, S.S.; Barrio, E.; Querol, A. Molecular characterization of new natural hybrids between Saccharomyces cerevisiae and Saccharomyces kudriavzevii from brewing. Appl. Environ. Microbiol. 2008, 74, 2314–2320. [Google Scholar] [CrossRef] [Green Version]
- Lopandić, K.; Gangl, H.; Wallner, E.; Tscheik, G.; Leitner, G.; Querol, A.; Borth, N.; Breitenbach, M.; Prillinger, H.; Tiefenbrunner, W. Genetically different wine yeasts isolated from Austrian vine-growing regions influence wine aroma differently and contain putative hybrids between Saccharomyces cerevisiae and Saccharomyces kudriavzevii. FEMS Yeast Res. 2007, 7, 953–965. [Google Scholar] [CrossRef] [Green Version]
- Arroyo-Lopez, F.N.; Perez-Traves, L.; Querol, A.; Barrio, E. Exclusion of Saccharomyces kudriavzevii from a wine model system mediated by Saccharomyces cerevisiae. Yeast 2011, 28, 423–435. [Google Scholar] [CrossRef]
- Alonso-del-Real, J.; Contreras-Ruiz, A.; Castiglioni, G.L.; Barrio, E.; Querol, A. The use of mixed populations of Saccharomyces cerevisiae and S. kudriavzevii to reduce ethanol content in wine: Limited aeration, inoculum proportions, and sequential inoculation. Front. Microbiol. 2017, 8, 2087. [Google Scholar] [CrossRef]
- Costantini, A.; Cravero, M.C.; Panero, L.; Bonello, F.; Vaudano, E.; Pulcini, L.; Garcia-Moruno, E. Wine fermentation performance of indigenous Saccharomyces cerevisiae and Saccharomyces paradoxus strains isolated in a Piedmont vineyard. Beverages 2021, 7, 30. [Google Scholar] [CrossRef]
- Cameleyre, M.; Lytra, G.; Barbe, J.C. Static headspace analysis using low-pressure gas chromatography and mass spectrometry, application to determining multiple partition coefficients: A practical tool for understanding red wine fruity volatile perception and the sensory impact of higher alcohols. Anal Chem. 2018, 90, 10812–10818. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.L.; Frost, S.; Ugliano, M.; Cantu, A.R.; Currie, B.L.; Anderson, M.; Chassy, A.W.; Vidal, S.; Diéval, J.-B.; Aagaard, O.; et al. Sulfur dioxide–oxygen consumption ratio reveals differences in bottled wine oxidation. Am. J. Enol. Vitic. 2016, 67, 449–459. [Google Scholar] [CrossRef]
- Ferreira, V.; de la Fuente, A.; Sáenz-Navajas, M.P. Wine aroma vectors and sensory attributes. In Managing Wine Quality; Elsevier: Amsterdam, The Netherlands, 2022; pp. 3–39. [Google Scholar] [CrossRef]
- Majdak, A.; Herjavec, S.; Orlíc, S.; Redžepovíc, S.; Miroševíc, N. Comparison of wine aroma compounds produced by Saccharomyces paradoxus and Saccharomyces cerevisiae strains. Food Technol. Biotechnol. 2002, 40, 103–109. [Google Scholar]
- Sampaio, J.P.; Gonçalves, P. Natural populations of Saccharomyces kudriavzevii in Portugal are associated with Oak bark and are sympatric with S. cerevisiae and S. paradoxus. App. Environ. Microbiol. 2008, 74, 2144–2152. [Google Scholar] [CrossRef] [Green Version]
- Stribny, J.; Gamero, A.; Pérez-Torrado, R.; Querol, A. Saccharomyces kudriavzevii and Saccharomyces uvarum differ from Saccharomyces cerevisiae during the production of aroma-active higher alcohols and acetate esters using their amino acidic precursors. Int. J. Food Microbiol. 2015, 205, 41–46. [Google Scholar] [CrossRef] [Green Version]
- González, S.S.; Gallo, L.; Climent, M.D.; Barrio, E.; Querol, A. Enological characterization of natural hybrids from Saccharomyces cerevisiae and S. kudriavzevii. Int. J. Food Microbiol. 2007, 116, 11–18. [Google Scholar] [CrossRef]
- Ortiz-Tovar, G.; Minebois, R.; Barrio, E.; Querol, A.; Pérez-Torrado, R. Aroma production and fermentation performance of S. cerevisiae × S. kudriavzevii natural hybrids under cold oenological conditions. Int. J. Food Microbiol. 2019, 297, 51–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stribny, J.; Querol, A.; Pérez-Torrado, R. Differences in enzymatic properties of the Saccharomyces kudriavzevii and Saccharomyces uvarum alcohol acetyltransferases and their impact on aroma-active compounds production. Front. Microbiol. 2016, 7, 897. [Google Scholar] [CrossRef] [Green Version]
- Pérez, D.; Denat, M.; Minebois, R.; Heras, J.M.; Guillamón, J.M.; Ferreira, V.; Querol, A. Modulation of aroma and chemical composition of Albariño semi-synthetic wines by non-wine Saccharomyces yeasts and bottle aging. Food Microbiol. 2022, 104, 103981. [Google Scholar] [CrossRef]



| Fermentation Time (dd) | pH | Total Acidity 1 (g/L) | °Babo | Alcoholic Degree (%v/v) | Yeast (Log CFU/mL) |
|---|---|---|---|---|---|
| 0 | 3.55 | 9.25 | 22.5 | 0.0 | 5.58 |
| 7 | 3.69 | 9.07 | 16.2 | 3.2 | 7.77 |
| 14 | 3.78 | 8.55 | 5.3 | 10.1 | 7.29 |
| 21 | 3.91 | 6.26 | 1.5 | 12.8 | 6.88 |
| 28 | 3.88 | 6.04 | 0.0 | 13.6 | 6.31 |
| Isolate | ITS (bp) | ITS-RFLP Patterns | Patterns | Species | RFLP Patterns of Nuclear Genes (Restriction Enzyme) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HaeIII | CfoI | HinfI | Interdelta | Dan4 | CAT8 (CfoI) | CYR1 (HaeIII) | GSY1 (MspI) | MET6 (HinfI) | OPY1 (HaeIII) | |||
| F0-51 | 380 | 280-120 | 200-100 | 1 nd | 2 na | na | 3M. pulcherrima | Na | na | na | na | na |
| F0-52 | 750 | 750 | 320-310-105 | nd | na | na | H. uvarum | Na | na | na | na | na |
| F7-61 | 380 | 280-120 | 200-100 | nd | na | na | M. pulcherrima | Na | na | na | na | na |
| F7-71 | 850 | 320-230-180-150 | 385-365-140 | 365-150 | A | A | S. cerevisiae | Nd | nd | nd | nd | nd |
| F7-72 | 850 | 500-230-150 | 385-365-140 | 365-150 | B | 4 nr | 1S. kudriavzevii | 560-175 | 295-155-80-30 | 340-270-160 | 650 | 520-300 |
| F7-73 | 750 | 750 | 320-310-105 | nd | na | na | H. uvarum | Nd | nd | nd | nd | nd |
| F7-74 | 850 | 320-230-180-150 | 385-365-140 | 365-150 | A | A | S. cerevisiae | Nd | nd | nd | nd | nd |
| F14-61 | 850 | 500-230-150 | 385-365-140 | 365-150 | B | nr | S. kudriavzevii | Nd | nd | nd | nd | nd |
| F14-62 | 850 | 320-230-180-150 | 385-365-140 | 365-150 | A | A | 1S. cerevisiae | 750 | 560 | 610-160 | nd | 750 |
| F14-63 | 850 | 320-230-180-150 | 385-365-140 | 365-150 | A | A | S. cerevisiae | Nd | nd | nd | nd | nd |
| F14-71 | 850 | 320-230-180-150 | 385-365-140 | 365-150 | A | A | S. cerevisiae | Nd | nd | nd | nd | nd |
| F21-61 | 850 | 320-230-180-150 | 385-365-140 | 365-150 | A | A | S. cerevisiae | Nd | nd | nd | nd | nd |
| F21-62 | 850 | 320-230-180-150 | 385-365-140 | 365-150 | C | nr | 1S. paradoxus | 750 | 400-210 | 420-350 | nd | 520-300 |
| F21-51 | 480 | 480 | 200-100-50 | nd | na | na | 1St. bacillaris | Na | na | na | na | na |
| F21-52 | 850 | 320-230-180-150 | 385-365-140 | 365-150 | A | A | S. cerevisiae | Nd | nd | nd | nd | nd |
| F28-51 | 850 | 320-230-180-150 | 385-365-140 | 365-150 | A | A | S. cerevisiae | Nd | nd | nd | nd | nd |
| F28-52 | 850 | 320-230-180-150 | 385-365-140 | 365-150 | C | nr | S. paradoxus | Nd | nd | nd | nd | nd |
| F28-53 | 850 | 320-230-180-150 | 385-365-140 | 365-150 | A | A | S. cerevisiae | Nd | nd | nd | nd | nd |
| F28-61 | 850 | 320-230-180-150 | 385-365-140 | 365-150 | A | A | S. cerevisiae | Nd | nd | nd | nd | nd |
| F28-62 | 850 | 320-230-180-150 | 385-365-140 | 365-150 | A | A | S. cerevisiae | Nd | nd | nd | nd | nd |
| EC1118 | nd | nd | nd | nd | nd | nd | S. cerevisiae | 750 | 560 | 610-160 | nd | 750 |
| Strain | Gene | Species | Closely Related Accession Number | Similarity |
|---|---|---|---|---|
| F0-51 | ITS (rDNA) | M. pulcherrima | NR_164379.1 | 100% |
| F0-52 | ITS (rDNA) | H. uvarum | NR_130660.1 | 99% |
| F21-51 | ITS (rDNA) | St. bacillaris | KY102528 | 100% |
| F14-62 | ITS (rDNA) | S. cerevisiae | NR_111007.1 | 98% |
| F7-72 | ITS (rDNA) | S. kudriavzevii | NR_111355.1 | 99% |
| F7-72 | CAT8 | S. kudriavzevii | LR215963.1 | 99% |
| F7-72 | CYR1 | S. kudriavzevii | LR215960.1 | 99% |
| F7-72 | MET6 | S. kudriavzevii | LR215939.1 | 99% |
| F7-72 | GSY1 | S. kudriavzevii | LR215952.1 | 99% |
| F7-72 | OPY1 | S. kudriavzevii | LR215952.1 | 99% |
| F21-62 | ITS (rDNA) | S. paradoxus | NR_138272.1 | 99% |
| F21-62 | CAT8 | S. paradoxus | XM_033912691.1 | 99% |
| F21-62 | CYR1 | S. paradoxus | XM_033911374.1 | 99% |
| F21-62 | GSY1 | S. paradoxus | XM_033910188.1 | 99% |
| F21-62 | OPY1 | S. paradoxus | XM_033908831.1 | 100% |
| Strain | pH | TA | VA | T-SO2 | F-SO2 | GF | Tac | Mac | Sac | GLY | ET |
|---|---|---|---|---|---|---|---|---|---|---|---|
| F7-72(Sk_B) | 2.96 ± 0.01 b | 6.60 ± 0.04 b | 0.31 ± 0.02 b | 3.18 ± 0.37 b | 0.64 ± 0.06 a | 1.53 ± 1.23 b | 4.87 ± 0.06 a | 1.10 ± 0.00 a | 2.20 ± 0.02 b | 5.43 ± 0.16 a | 10.70 ± 0.89 c |
| F14-62(Sc_A) | 2.99 ± 0.03 b | 7.01 ± 0.09 a | 0.55 ± 0.01 a | 14.66 ± 0.041 a | 0.82 ± 0.09 a | 0.08 ± 0.05 b | 5.00 ± 0.03 a | 1.16 ±0.03 a | 1.980.17 b | 4.30 ± 0.09 b | 13.53 ± 0.11 a |
| F21-62(Sp_C) | 2.96 ± 0.02 b | 6.93 ± 0.16 a | 0.42 ± 0.01 ab | 3.15 ± 0.05 b | 0.74 ± 0.20 a | 4.53 ± 0.24 a | 4.86 ± 0.29 a | 1.13 ± 0.06 a | 1.22 0.78 a | 5.54 ± 0.20 a | 12.59 ± 0.18 b |
| RT | Compounds | F7-72 (µg/L) | F14-62 (µg/L) | F21-62 (µg/L) | |||
|---|---|---|---|---|---|---|---|
| Esters | |||||||
| 15.072 | Ethyl butyrate | 176.24 ± 20.53 | a | 92.79 ± 5.98 | b | 174.65 ± 11.50 | a |
| 17.070 | Butyl acetate | 2.11 ± 0.11 | ns | 2.82 ± 0.10 | ns | 2.61 ± 1.28 | ns |
| 20.093 | Isoamyl acetate | 226.15 ± 13.72 | b | 408.38 ± 8.41 | a | 249.70 ± 15.92 | b |
| 27.420 | Ethyl hexanoate | 331.46 ± 29.74 | a | 245.01 ± 23.79 | b | 318.38 ± 10.60 | a |
| 30.021 | Hexyl acetate | 15.47 ± 1.90 | b | 25.51 ± 0.51 | a | 21.88 ± 2.11 | a |
| 34.791 | Ethyl lactate | 36.66 ± 6.88 | ns | 30.48 ± 2.27 | ns | 39.34 ± 4.44 | ns |
| 40.827 | Ethyl octanoate | 168.64 ± 6.98 | ns | 132.49 ± 67.49 | ns | 158.36 ± 32.75 | ns |
| 46.237 | Ethyl 3-hydroxybutyrate | 7.37 ± 1.75 | ns | 3.78 ± 0.42 | ns | 7.14 ± 1.01 | ns |
| 53.417 | Ethyl decanoate | 47.42 ± 1.75 | a | 3.35 ± 0.21 | b | 33.67 ± 15.17 | ab |
| 55.624 | Diethyl succinate | 86.97 ± 14.32 | a | 30.71 ± 2.25 | b | 42.59 ± 3.00 | b |
| 56.451 | Ethyl 9-decenoate | 52.69 ± 0.11 | ns | 75.67 ± 64.65 | ns | 59.77 ± 21.83 | ns |
| 59.261 | Ethyl acetate | 18.69 ± 3.23 | ns | 13.86 ± 0.86 | ns | 17.65 ± 0.44 | ns |
| 63.347 | beta-Phenethyl acetate | 114.46 ± 4.54 | a | 19.56 ± 0.83 | c | 53.02 ± 2.33 | b |
| 67.500 | Ethyl 3-hydroxyhexanoate | 2.72 ± 0.65 | b | 6.88 ± 0.09 | a | 3.20 ± 1.15 | b |
| 90.526 | Ethyl hydrogen succinate | 74.97 ± 6.18 | ns | 70.37 ± 30.17 | ns | 50.17 ± 4.85 | ns |
| Tot Esters | 1362.02 ± 55.67 | ns | 1161.65 ± 108.93 | ns | 1232.13 ± 22.96 | ns | |
| Alcohols | |||||||
| 18.187 | Isobutyl alcohol | 303.62 ± 20.18 | b | 705.87 ± 26.73 | a | 192.20 ± 26.81 | b |
| 21.581 | 1-Butanol | 31.18 ± 2.17 | b | 4.83 ± 0.41 | c | 67.99 ± 9.71 | a |
| 25.962 | 3- + 2-methyl-1-butanol | 10,771.47 ± 307.35 | ab | 13,128.03 ± 454.11 | a | 10,162.02 ± 817.31 | b |
| 33.910 | 3-methyl-1-pentanol | 13.48 ± 1.88 | b | 6.08 ± 0.77 | c | 20.62 ± 1.26 | a |
| 35.670 | 1-Hexanol | 733.39 ± 11.72 | ns | 737.89 ± 14.98 | ns | 792.96 ± 63.41 | ns |
| 36.315 | (E)-3-hexen-1-ol | 4.48 ± 0.85 | ns | 5.83 ± 1.13 | ns | 5.40 ± 0.41 | ns |
| 37.671 | (Z)-3-hexen-1-ol | 22.99 ± 0.28 | ns | 20.02 ± 0.40 | ns | 20.26 ± 1.29 | ns |
| 42.352 | 1-heptanol | 26.81 ± 1.58 | b | 38.17 ± 0.93 | a | 32.49 ± 1.96 | ab |
| 44.510 | 2-ethyl-1-hexanol | 5.58 ± 1.14 | ns | 5.06 ± 0.65 | ns | 5.70 ± 1.40 | ns |
| 48.764 | 1-Octanol | 6.59 ± 0.51 | a | 1.28 ± 0.78 | c | 3.56 ± 0.60 | b |
| 68.579 | Phenylethyl Alcohol | 7802.38 ± 224.25 | a | 4023.73 ± 44.05 | b | 7102.09 ± 372.12 | a |
| Tot Alcohols | 19,721.96 ± 115.92 | ns | 18,676.77 ± 527.82 | ns | 18,405.28 ± 1173.81 | ns | |
| Acids | |||||||
| 41.747 | Acetic acid | 271.48 ± 184.28 | ns | 303.22 ± 27.87 | ns | 227.73 ± 32.15 | ns |
| 49.165 | Isobutyric acid | 18.60 ± 0.93 | a | 8.96 ± 1.47 | b | 5.68 ± 0.62 | b |
| 52.765 | Butanoic acid | 2.81 ± 0.46 | ab | 0.78 ± 0.40 | b | 3.91 ± 1.54 | a |
| 55.284 | Isovaleric acid | 43.81 ± 7.99 | ns | 31.42 ± 9.61 | ns | 52.93 ± 9.05 | ns |
| 64.966 | Hexanoic acid | 353.46 ± 18.34 | ns | 241.87 ± 66.07 | ns | 328.66 ± 59.87 | ns |
| 75.863 | Octanoic Acid | 1169.95 ± 63.35 | ns | 807.08 ± 127.31 | ns | 717.04 ± 127.28 | ns |
| 80.932 | Nonanoic acid | 5.16 ± 0.23 | a | 0.00 ± 0.00 | b | 3.81 ± 1.21 | a |
| 85.752 | n-Decanoic acid | 452.59 ± 47.03 | a | 15.66 ± 8.13 | b | 388.58 ± 25.67 | a |
| 88.443 | 9-Decenoic acid | 443.78 ± 55.42 | ns | 588.22 ± 91.65 | ns | 547.06 ± 78.08 | ns |
| Tot Acids | 2761.64 ± 323.98 | ns | 1997.21 ± 299.11 | ns | 2275.40 ± 181.07 | ns | |
| Miscellaneous | |||||||
| 12.166 | 2-Pentanone | 7.73 ± 1.61 | a | 8.03 ± 0.44 | a | 3.45 ± 0.43 | b |
| 30.739 | Acetoin | 87.82 ± 10.32 | a | 25.28 ± 0.73 | b | 39.21 ± 2.33 | b |
| 52.572 | Butyrolactone | 30.49 ± 4.63 | b | 41.28 ± 0.57 | a | 33.16 ± 2.07 | b |
| 56.949 | p-menth-1-en-8-ol | 4.99 ± 1.07 | ns | 5.25 ± 0.73 | ns | 5.80 ± 0.36 | ns |
| 58.048 | 3-(methylthio)-1-propanol | 25.95 ± 0.22 | b | 37.08 ± 1.51 | a | 21.16 ± 1.68 | b |
| 60.817 | beta-citronellol | 5.96 ± 0.81 | ab | 7.21 ± 0.45 | a | 5.42 ± 0.45 | b |
| 70.661 | Benzothiazole | 7.93 ± 0.41 | ab | 14.76 ± 4.72 | a | 0.00 ± 0.00 | b |
| 82.159 | 4-vinyl guaiacol | 25.87 ± 7.14 | a | 2.38 ± 0.39 | b | 39.98 ± 1.29 | a |
| Tot Miscellaneous | 196.74 ± 6.66 | a | 141.26 ± 5.04 | b | 148.19 ± 4.86 | b | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petruzziello, E.; Blaiotta, G.; Pittari, E.; Piombino, P.; Aponte, M. Isolation and Characterization of Cryotolerant Yeasts from Fiano di Avellino Grapes Fermented at Low Temperatures. Foods 2023, 12, 526. https://0-doi-org.brum.beds.ac.uk/10.3390/foods12030526
Petruzziello E, Blaiotta G, Pittari E, Piombino P, Aponte M. Isolation and Characterization of Cryotolerant Yeasts from Fiano di Avellino Grapes Fermented at Low Temperatures. Foods. 2023; 12(3):526. https://0-doi-org.brum.beds.ac.uk/10.3390/foods12030526
Chicago/Turabian StylePetruzziello, Ernesto, Giuseppe Blaiotta, Elisabetta Pittari, Paola Piombino, and Maria Aponte. 2023. "Isolation and Characterization of Cryotolerant Yeasts from Fiano di Avellino Grapes Fermented at Low Temperatures" Foods 12, no. 3: 526. https://0-doi-org.brum.beds.ac.uk/10.3390/foods12030526