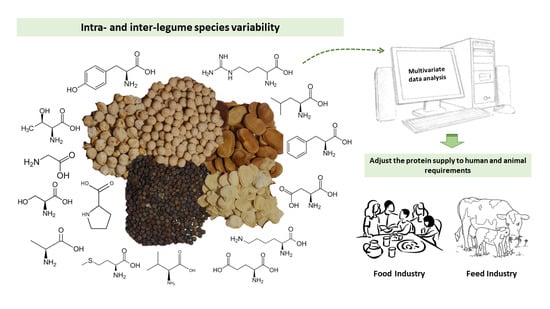
High Inter- and Intra- Diversity of Amino Acid Content and Protein Digestibility Disclosed in Five Cool Season Legume Species with a Growing Market Demand
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Chemicals and Reagents
2.3. Samples Preparation
2.4. Total Protein
2.5. Protein Quality
2.5.1. Amino Acids’ Extraction
2.5.2. Amino Acids’ Content
2.5.3. In vitro Protein Digestibility
2.5.4. Calculated Protein Quality
2.6. Statistical Analyses
3. Results and Discussion
3.1. High Diversity Detected in Protein Content and Quality among but Also within Five Cool Season Grain Legume Species
3.2. Integrating the Protein Diversity in the Legume World—Featuring the Differences
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vaz Patto, M.C. Grain legume protein quality: A hot subject. Arbor 2016, 192, a314. [Google Scholar] [CrossRef]
- United Nations. UN Department of Economic and Social Affairs, Population Dynamics. World Population Prospects 2022. 2022. Available online: https://population.un.org/wpp/ (accessed on 25 July 2022).
- Semba, R.D.; Ramsing, R.; Rahman, N.; Kraemer, K.; Bloem, M.W. Legumes as a sustainable source of protein in human diets. Glob. Food Sec. 2021, 28, 100520. [Google Scholar] [CrossRef]
- Stagnari, F.; Maggio, A.; Galieni, A.; Pisante, M. Multiple benefits of legumes for agriculture sustainability: An overview. Chem. Biol. Technol. Agric. 2017, 4, 2. [Google Scholar] [CrossRef]
- Rubiales, D.; Annicchiarico, P.; Vaz Patto, M.C.; Julier, B. Legume breeding for the agroecological transition of global agri-food systems: A European perspective. Front. Plant Sci. 2021, 12, 782574. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, L.; Rubiales, D.; Bronze, M.R.; Vaz Patto, M.C. Grass pea (Lathyrus sativus L.)—A sustainable and resilient answer to climate challenges. Agronomy 2022, 12, 1324. [Google Scholar] [CrossRef]
- Tor-Roca, A.; Garcia-Aloy, M.; Mattivi, F.; Llorach, R.; Andres-Lacueva, C.; Urpi-Sarda, M. Phytochemicals in legumes: A qualitative reviewed analysis. J. Agric. Food Chem. 2020, 68, 13486–13496. [Google Scholar] [CrossRef]
- Lee, H.J.; Yong, H.I.; Kim, M.; Choi, Y.-S.; Jo, C. Status of meat alternatives and their potential role in the future meat market—A review. Asian-Australas J. Anim. Sci. 2020, 33, 1533–1543. [Google Scholar] [CrossRef]
- Elamine, Y.; Alaiz, M.; Girón-Calle, J.; Guiné, R.P.F.; Vioque, J. Nutritional characteristics of the seed protein in 23 Mediterranean legumes. Agronomy 2022, 12, 400. [Google Scholar] [CrossRef]
- Miles, R.D.; Chapman, F.A. The Concept of Ideal Protein in Formulation of Aquaculture Feeds. School of Forest Resources and Conservation, Program in Fisheries and Aquatic Sciences, UF/IFAS Extension, FA144, 1–4. Available online: https://edis.ifas.ufl.edu/publication/FA144 (accessed on 25 August 2022).
- Szepe, K.J.; Dyer, P.S.; Johnson, R.I.; Salter, A.M.; Avery, S.V. Influence of environmental and genetic factors on food protein quality: Current knowledge and future directions. Curr. Opin. Food Sci. 2021, 40, 94–101. [Google Scholar] [CrossRef]
- Food DataCentral USDA. Available online: https://fdc.nal.usda.gov/ (accessed on 20 October 2022).
- Serrano, C.; Carbas, B.; Castanho, A.; Soares, A.; Vaz Patto, M.C.; Brites, C. Characterisation of nutritional quality traits of a chickpea (Cicer arietinum) germplasm collection exploited in chickpea breeding in Europe. Crop Pasture Sci. 2017, 68, 1031–1040. [Google Scholar] [CrossRef]
- Santos, C.S.; Carbas, B.; Castanho, A.; Bronze, M.R.; Serrano, C.; Vasconcelos, M.W.; Vaz Patto, M.C.; Brites, C. Relationship between seed traits and pasting and cooking behaviour in a pulse germplasm collection. Crop Pasture Sci. 2018, 69, 892–903. [Google Scholar] [CrossRef]
- Santos, C.S.; Carbas, B.; Castanho, A.; Vasconcelos, M.W.; Vaz Patto, M.C.; Domoney, C.; Brites, C. Variation in pea (Pisum sativum L.) seed quality traits defined by physicochemical functional properties. Foods 2019, 8, 570. [Google Scholar] [CrossRef]
- ISO 16634; Food Products—Determination of the Total Nitrogen Content by Combustion According to Dumas’ Principle and Calculation of the Crude Protein Content. International Organization for Standardization: London, UK, 2016.
- Igor, J.; Krstović, S.; Glamocic, D.; Jakšić, S.; Abramović, B. Validation of an HPLC method for the determination of amino acids in feed. J. Serb. Chem. Soc. 2013, 78, 839–850. [Google Scholar] [CrossRef]
- Mecha, E.; Natalello, S.; Carbas, B.; da Silva, A.B.; Leitão, S.T.; Brites, C.; Veloso, M.M.; Rubiales, D.; Costa, J.; Cabral, M.d.F.; et al. Disclosing the nutritional quality diversity of Portuguese common beans—The missing link for their effective use in protein quality breeding programs. Agronomy 2021, 11, 221. [Google Scholar] [CrossRef]
- Tinus, T.; Damour, M.; van Riel, V.; Sopade, P.A. Particle size-starch–protein digestibility relationships in cowpea (Vigna unguiculata). J. Food Eng. 2012, 113, 254–264. [Google Scholar] [CrossRef]
- Young, V.R.; Pellett, P.L. Plant proteins in relation to human protein and amino acid nutrition. Am. J. Clin. Nutr. 1994, 59, 1203S–1212S. [Google Scholar] [CrossRef]
- FAO. Dietary Protein Quality Evaluation in Human Nutrition. Available online: https://www.fao.org/ag/humannutrition/35978-02317b979a686a57aa4593304ffc17f06.pdf (accessed on 25 July 2022).
- Pastor-Cavada, E.; Juan, R.; Pastor, J.E.; Alaiz, M.; Vioque, J. Protein and amino acid composition of select wild legume species of tribe Fabeae. Food Chem. 2014, 163, 97–102. [Google Scholar] [CrossRef]
- Payne, R.; Harding, S. GenStat for Windows. Introduction, 18th ed.; VSN International: Hemel Hempstead, UK, 2015. [Google Scholar]
- Rathi, D.; Chakraborty, S.; Chakraborty, N. Grasspea, a critical recruit among neglected and underutilized legumes, for tapping genomic resources. Curr. Plant Biol. 2021, 26, 100200. [Google Scholar] [CrossRef]
- Kumari, S.; Jha, V.K.; Kumari, D.; Ranjan, R.; Nimmy, M.S.; Kumar, A.; Kishore, C.; Kumar, V. Protein content of Lathyrus sativus collected from diverse locations. J. Pharmacogn. Phytochem. 2018, 7, 1610–1611. [Google Scholar]
- Ramya, K.R.; Tripathi, K.; Pandey, A.; Barpete, S.; Gore, P.G.; Raina, A.P.; Khawar, K.M.; Swain, N.; Sarker, A. Rediscovering the potential of multifaceted orphan legume grasspea—A sustainable resource with high nutritional values. Front. Nutr. 2022, 8, 2021. [Google Scholar] [CrossRef]
- Khazaei, H.; Subedi, M.; Nickerson, M.; Martínez-Villaluenga, C.; Frias, J.; Vandenberg, A. Seed protein of lentils: Current status, progress, and food applications. Foods. 2019, 8, 391. [Google Scholar] [CrossRef] [PubMed]
- Hang, J.; Shi, D.; Neufeld, J.; Bett, K.E.; House, J.D. Prediction of protein and amino acid contents in whole and ground lentils using near-infrared reflectance spectroscopy. LWT 2022, 165, 113669. [Google Scholar] [CrossRef]
- Ciurescu, G.; Toncea, I.; Mariana, R.; Habeanu, M. Seeds composition and their nutrients quality of some pea (Pisum sativum L.) and lentil (Lens culinaris medik.) cultivars. Rom. Agric. Res. 2018, 2018, 101–108. [Google Scholar]
- Grasso, N.; Lynch, N.L.; Arendt, E.K.; O’Mahony, J.A. Chickpea protein ingredients: A review of composition, functionality, and applications. Compr. Rev. Food Sci. Food Saf. 2022, 21, 435–452. [Google Scholar] [CrossRef] [PubMed]
- Sandeep Kaur, D.; Pooja, S.; Prashant, K. Pea seed proteins: A nutritional and nutraceutical Update. In Grain and Seed Proteins Functionality, 1st ed.; Jimenez-Lopez, J.C., Ed.; IntechOpen: Rijeka, Croatia, 2021; pp. 1–16. [Google Scholar]
- Taghvaei, M.; Sadeghi, R.; Smith, B. Seed to seed variation of proteins of the yellow pea (Pisum sativum L.). PLoS ONE 2022, 17, e0271887. [Google Scholar] [CrossRef]
- Hood-Niefer, S.D.; Warkentin, T.D.; Chibbar, R.N.; Vandenberg, A.; Tyler, R.T. Effect of genotype and environment on the concentrations of starch and protein in, and the physicochemical properties of starch from, field pea and faba bean. J. Sci. Food Agric. 2012, 92, 141–150. [Google Scholar] [CrossRef]
- Meng, Z.; Liu, Q.; Zhang, Y.; Chen, J.; Sun, Z.; Ren, C.; Zhang, Z.; Cheng, X.; Huang, Y. Nutritive value of faba bean (Vicia faba L.) as a feedstuff resource in livestock nutrition: A review. Food Sci. Nutr. 2021, 9, 5244–5262. [Google Scholar] [CrossRef]
- Mayer Labba, I.-C.; Frøkiær, H.; Sandberg, A.-S. Nutritional and antinutritional composition of fava bean (Vicia faba L., var. minor) cultivars. Food Res. Int. 2021, 140, 110038. [Google Scholar] [CrossRef]
- Wu, G.; Bazer, F.W.; Davis, T.A.; Kim, S.W.; Li, P.; Marc Rhoads, J.; Carey Satterfield, M.; Smith, S.B.; Spencer, T.E.; Yin, Y. Arginine metabolism and nutrition in growth, health and disease. Amino Acids 2009, 37, 153–168. [Google Scholar] [CrossRef]
- Platt, S.R. The role of glutamate in central nervous system health and disease—A review. Vet. J. 2007, 173, 278–286. [Google Scholar] [CrossRef]
- Brosnan, J.T.; Brosnan, M.E. Glutamate: A truly functional amino acid. Amino Acids 2013, 45, 413–418. [Google Scholar] [CrossRef]
- Karetnikova, E.S.; Jarzebska, N.; Markov, A.G.; Weiss, N.; Lentz, S.R.; Rodionov, R.N. Is homoarginine a protective cardiovascular risk factor? Arter. Thromb. Vasc. Biol. 2019, 39, 869–875. [Google Scholar] [CrossRef]
- Mendes, F.A.; Leitão, S.T.; Correia, V.; Mecha, E.; Rubiales, D.; Bronze, M.R.; Vaz Patto, M.C. Portuguese common bean natural variation helps to clarify the genetic architecture of the legume’s nutritional composition and protein quality. Plants 2021, 11, 26. [Google Scholar] [CrossRef]
- Qiu, X.-M.; Sun, Y.-Y.; Ye, X.-Y.; Li, Z.-G. Signaling role of glutamate in plants. Front. Plant Sci. 2020, 10, 1743. [Google Scholar] [CrossRef]
- Winter, G.; Todd, C.D.; Trovato, M.; Forlani, G.; Funck, D. Physiological implications of arginine metabolism in plants. Front. Plant Sci. 2015, 6, 534. [Google Scholar] [CrossRef]
- Kumar, V.; Sharma, A.; Kaur, R.; Thukral, A.K.; Bhardwaj, R.; Ahmad, P. Differential distribution of amino acids in plants. Amino Acids 2017, 49, 821–869. [Google Scholar] [CrossRef]
- Liu, G.; Zhang, M.; Chen, X.; Zhang, W.; Ding, W.; Zhang, Q. Evolution of threonine aldolases, a diverse family involved in the second pathway of glycine biosynthesis. J. Mol. Evol. 2015, 80, 102–107. [Google Scholar] [CrossRef]
- Yang, Q.; Zhao, D.; Liu, Q. Connections between amino acid metabolisms in plants: Lysine as an example. Front. Plant Sci. 2020, 11, 928. [Google Scholar] [CrossRef]
- Ingle, R.A. Histidine biosynthesis. In The Arabidopsis Book; American Society of Plant Biologists: Rockville, MD, USA, 2011. [Google Scholar] [CrossRef]
- Bromke, M.A. Amino acid biosynthesis pathways in diatoms. Metabolites 2013, 3, 294–311. [Google Scholar] [CrossRef]
- Matemu, A.; Nakamura, S.; Katayama, S. Health benefits of antioxidative peptides derived from legume proteins with a high amino acid score. Antioxidants 2021, 10, 316. [Google Scholar] [CrossRef]
- Apone, F.; Barbulova, A.; Colucci, M.G. Plant and microalgae derived peptides are advantageously employed as bioactive compounds in cosmetics. Front. Plant Sci. 2019, 10, 756. [Google Scholar] [CrossRef] [PubMed]
- Indrati, R. Bioactive peptides from legumes and their bioavailability. In Legumes Research, 1st ed.; Jimenez-Lopez, J.C., Clemente, A., Eds.; IntechOpen: London, UK, 2021; Volume 2, pp. 1–27. [Google Scholar] [CrossRef]
- Sánchez-Velázquez, O.A.; Ribéreau, S.; Mondor, M.; Cuevas-Rodríguez, E.O.; Arcand, Y.; Hernández-Álvarez, A.J. Impact of processing on the in vitro protein quality, bioactive compounds, and antioxidant potential of 10 selected pulses. Legume Sci. 2021, 3, e88. [Google Scholar] [CrossRef]
- Mendes, F.; De Almeida Oliveira, M.G.; Costa, N.M.B.; Pires, C.V.; Passos, F.R. Capability of in vitro digestibility methods to predict in vivo digestibility of vegetal and animal proteins. Arch. Latinoam. Nutr. 2016, 66, 5–16. [Google Scholar]
- Chernukha, I.; Meliashchenia, A.; Kaltovich, I.V.; Vasilevskaya, E.; Aryzina, M.; Smaliak, T.; Senchenko, T.; Fedulova, L. Evolution of in vitro digestibility techniques: A systematic review. Theory Pract. Meat Process. 2022, 6, 300–310. [Google Scholar] [CrossRef]
- Ohanenye, I.C.; Ekezie, F.-G.C.; Sarteshnizi, R.A.; Boachie, R.T.; Emenike, C.U.; Sun, X.; Nwachukwu, I.D.; Udenigwe, C.C. Legume seed protein digestibility as influenced by traditional and emerging physical processing technologies. Foods 2022, 11, 2299. [Google Scholar] [CrossRef]
- Sim, S.Y.J.; Srv, A.; Chiang, J.H.; Henry, C.J. Plant proteins for future foods: A roadmap. Foods 2021, 10, 1967. [Google Scholar] [CrossRef]
- Gu, J.; Bk, A.; Wu, H.; Lu, P.; Nawaz, M.A.; Barrow, C.J.; Dunshea, F.R.; Suleria, H.A.R. Impact of processing and storage on protein digestibility and bioavailability of legumes. Food Rev. Int. 2022, 1–28. [Google Scholar] [CrossRef]
- Davies, A.M.C.; Fearn, T. Back to Basics: Multivariate Quantitative Analysis, Canonical Variate Analysis. 2008. Available online: www.spectroscopyeurope.com (accessed on 29 August 2022).




| CA (n = 86) | PS (n = 118) | VF (n = 92) | LC (n = 92) | LS (n = 109) | |
|---|---|---|---|---|---|
| AAS_Val | 143.85 ± 14.41 b | 112.04 ± 22.41 d | 123.13 ± 12.97 c | 165.87 ± 21.59 a | 117.12 ± 12.60 d |
| AAS_Thr | 96.43± 17.56 a | 38.46± 8.62 d | 75.96± 12.37 b | 61.76± 14.12 c | 35.33± 19.04 d |
| AAS_Ile | 168.07 ± 18.20 a | 100.62 ± 19.22 d | 129.99 ± 11.20 c | 142.19 ± 17.89 b | 127.02 ± 14.08 c |
| AAS_Leu | 114.96 ± 9.96 a | 81.64 ± 14.04 e | 94.97 ± 7.66 c | 109.63 ± 14.32 b | 89.47 ± 8.42 d |
| AAS_Met | 32.37 ± 3.48 a | 17.09 ± 2.25 cd | 17.64 ± 1.75 c | 20.61 ± 3.40 b | 16.37 ± 1.89 d |
| AAS_His | 119.56 ± 9.84 a | 57.52 ± 9.47 c | 101.12 ± 8.69 b | 119.05 ± 16.08 a | 99.72 ± 11.23 b |
| AAS_Phe + Tyr | 127.00 ± 10.35 a | 89.55 ± 17.12 c | 92.51 ± 7.53 c | 112.15 ± 13.46 b | 93.93 ± 9.57 c |
| AAS_Lys | 115.41 ± 9.98 b | 106.00 ± 15.52 c | 94.68 ± 8.62 d | 126.04 ± 17.17 a | 93.31 ± 8.82 d |
| PER1 | 2.59 ± 0.28 a | 1.62 ± 0.39 e | 2.02 ± 0.22 c | 2.42 ± 0.41 b | 1.85 ± 0.24 d |
| PER2 | 2.69 ± 0.28 a | 1.74 ± 0.38 e | 2.12 ± 0.21 c | 2.53 ± 0.40 b | 1.97 ± 0.23 d |
| PER3 | 2.39 ± 0.38 a | 0.69 ± 0.44 e | 1.36 ± 0.28 c | 1.96 ± 0.54 b | 1.18 ± 0.28 d |
| IVPD (%) | 71.04± 0.38 ab | 72.14± 0.31 ab | 70.54± 1.19 b | 73.13± 0.14 a | 71.91± 0.12 ab |
| IVPDCAAS | 22.48± 0.98 a | 12.35± 0.24 c | 11.16± 0.96 c | 16.29± 0.67 b | 11.93± 0.50 c |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mecha, E.; Alves, M.L.; Bento da Silva, A.; Pereira, A.B.; Rubiales, D.; Vaz Patto, M.C.; Bronze, M.R. High Inter- and Intra- Diversity of Amino Acid Content and Protein Digestibility Disclosed in Five Cool Season Legume Species with a Growing Market Demand. Foods 2023, 12, 1383. https://0-doi-org.brum.beds.ac.uk/10.3390/foods12071383
Mecha E, Alves ML, Bento da Silva A, Pereira AB, Rubiales D, Vaz Patto MC, Bronze MR. High Inter- and Intra- Diversity of Amino Acid Content and Protein Digestibility Disclosed in Five Cool Season Legume Species with a Growing Market Demand. Foods. 2023; 12(7):1383. https://0-doi-org.brum.beds.ac.uk/10.3390/foods12071383
Chicago/Turabian StyleMecha, Elsa, Mara Lisa Alves, Andreia Bento da Silva, Ana Bárbara Pereira, Diego Rubiales, Maria Carlota Vaz Patto, and Maria Rosário Bronze. 2023. "High Inter- and Intra- Diversity of Amino Acid Content and Protein Digestibility Disclosed in Five Cool Season Legume Species with a Growing Market Demand" Foods 12, no. 7: 1383. https://0-doi-org.brum.beds.ac.uk/10.3390/foods12071383