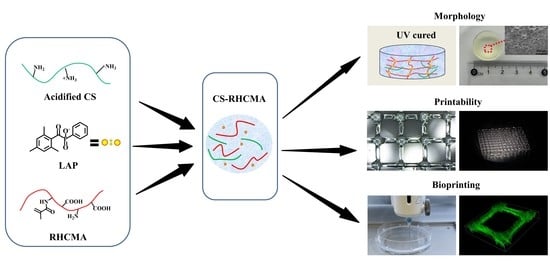
Preparation of Chitosan/Recombinant Human Collagen-Based Photo-Responsive Bioinks for 3D Bioprinting
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of the CS-RHCMA Bioinks
2.3. FT-IR Characterization
2.4. Morphological Characterization
2.5. Mechanical Testing
2.6. Biodegradation
2.7. Viscosity and Printability Test
2.8. Cell Culture
2.9. Cytotoxicity Test
2.10. 3D Bioprinted Constructs of HUVECs-Laden CS-RHCMA Bioinks
2.11. Statistical Analysis
3. Results and Discussion
3.1. Characterization of the CS-RHCMA
3.2. Internal Morphology
3.3. Mechanical Properties and Biodegradation
3.4. Viscosity and Printability
3.5. Cytotoxicity Test
3.6. 3D Bioprinting HUVECs-Laden CS-RHCMA Bioinks
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jorgensen, C.; Simon, M. In Vitro Human Joint Models Combining Advanced 3D Cell Culture and Cutting-Edge 3D Bioprinting Technologies. Cells 2021, 10, 596. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Tian, X.; Fan, J.; Tong, H.; Ao, Q.; Wang, X. An Interpenetrating Alginate/Gelatin Network for Three-Dimensional (3D) Cell Cultures and Organ Bioprinting. Molecules 2020, 25, 756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santoni, S.; Gugliandolo, S.G.; Sponchioni, M.; Moscatelli, D.; Colosimo, B.M. 3D Bioprinting: Current Status and Trends—A Guide to the Literature and Industrial Practice. Bio-Design Manuf. 2022, 5, 14–42. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Y. Tissue Engineering Applications of Three-Dimensional Bioprinting. Cell Biochem. Biophys. 2015, 72, 777–782. [Google Scholar] [CrossRef]
- Blaeser, A.; Campos, D.F.D.; Puster, U.; Richtering, W.; Stevens, M.M.; Fischer, H. Controlling Shear Stress in 3D Bioprinting is a Key Factor to Balance Printing Resolution and Stem Cell Integrity. Adv. Health Mater. 2016, 5, 326–333. [Google Scholar] [CrossRef]
- Li, Z.; Li, H.; Zhu, X.; Peng, Z.; Zhang, G.; Yang, J.; Wang, F.; Zhang, Y.; Sun, L.; Wang, R.; et al. Directly Printed Embedded Metal Mesh for Flexible Transparent Electrode via Liquid Substrate Electric-Field-Driven Jet. Adv. Sci. 2022, 9, 2105331. [Google Scholar] [CrossRef]
- Rommel, D.; Mork, M. Functionalized Microgel Rods Interlinked into Soft Macroporous Structures for 3D Cell Culture. Adv. Sci. 2022, 9, 2103554. [Google Scholar] [CrossRef]
- Terrell, J.A.; Jones, C.G.; Kabandana, G.K.M.; Chen, C. From Cells-On-A-Chip to Organs-on-a-Chip: Scaffolding Materials for 3D Cell Culture in Microfluidics. J. Mater. Chem. B 2020, 8, 6667–6685. [Google Scholar] [CrossRef]
- Chang, P.H.; Chao, H.M.; Chern, E.; Hsu, S.H. Chitosan 3D Cell Culture System Promotes Nave-like Features of Human Induced Pluripotent Stem Cells: A Novel Tool to Sustain Pluripotency and Facilitate Differentiation. Biomaterials 2021, 268, 120575. [Google Scholar] [CrossRef]
- Askari, M.; Naniz, M.A.; Kouhi, M.; Saberi, A.; Zolfagharian, A.; Bodaghi, M. Recent Progress in Extrusion 3D Bioprinting of Hydrogel Biomaterials for Tissue Regeneration: A Comprehensive Review with Focus on Advanced Fabrication Techniques. Biomater. Sci. 2021, 9, 535–573. [Google Scholar] [CrossRef]
- Jia, W.; Gungor-Ozkerim, P.S.; Zhang, Y.S.; Yue, K.; Zhu, K.; Liu, W.; Pi, Q.; Byambaa, B.; Dokmeci, M.R.; Shin, S.R.; et al. Direct 3D Bioprinting of Perfusable Vascular Constructs Using a Blend Bioink. Biomaterials 2016, 106, 58–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, Z.; Wang, J.; Tian, J.; Deng, X.; Balayan, A.; Sun, Y.; Xiang, Y.; Guan, J.; Schimelman, J.; Hwang, H.; et al. Rapid 3D Bioprinting of a Multicellular Model Recapitulating Pterygium Microenvironment. Biomaterials 2022, 282, 121391. [Google Scholar] [CrossRef] [PubMed]
- Lelièvre, S.A.; Kwok, T.; Chittiboyina, S. Architecture in 3D Cell Culture: An Essential Feature for in Vitro Toxicology. Toxicol. Vitr. 2017, 45, 287–295. [Google Scholar] [CrossRef] [PubMed]
- You, F.; Wu, X.; Kelly, M.; Chen, X. Bioprinting and in Vitro Characterization of Alginate Dialdehyde–Gelatin Hydrogel Bio-Ink. Bio-Design Manuf. 2020, 3, 48–59. [Google Scholar] [CrossRef] [Green Version]
- Tytgat, L.; Dobos, A.; Markovic, M.; Van Damme, L.; Van Hoorick, J.; Bray, F.; Thienpont, H.; Ottevaere, H.; Dubruel, P.; Ovsianikov, A.; et al. High-Resolution 3D Bioprinting of Photo-Cross-linkable Recombinant Collagen to Serve Tissue Engineering Applications. Biomacromolecules 2020, 21, 3997–4007. [Google Scholar] [CrossRef]
- Roehm, K.D.; Madihally, S.V. Bioprinted Chitosan-Gelatin Thermosensitive Hydrogels Using an Inexpensive 3D Printer. Biofabrication 2017, 10, 015002. [Google Scholar] [CrossRef]
- Weng, T.; Zhang, W.; Xia, Y.; Wu, P.; Yang, M.; Jin, R.; Xia, S.; Wang, J.; You, C.; Han, C.; et al. 3D Bioprinting for Skin Tissue Engineering: Current Status and Perspectives. J. Tissue Eng. 2021, 12, 20417314211028574. [Google Scholar] [CrossRef]
- Roque, R.; Barbosa, G.F.; Guastaldi, A.C. Design and 3D Bioprinting of Interconnected Porous Scaffolds for Bone Regeneration. An Additive Manufacturing Approach. J. Manuf. Process. 2021, 64, 655–663. [Google Scholar] [CrossRef]
- Abbadessa, A.; Mouser, V.H.M.; Blokzijl, M.M.; Gawlitta, D.; Dhert, W.J.A.; Hennink, W.E.; Malda, J.; Vermonden, T. A Synthetic Thermosensitive Hydrogel for Cartilage Bioprinting and Its Biofunctionalization with Polysaccharides. Biomacromolecules 2016, 17, 2137–2147. [Google Scholar] [CrossRef] [Green Version]
- Xing, F.; Xiang, Z.; Rommens, P.M.; Ritz, U. 3D Bioprinting for Vascularized Tissue-Engineered Bone Fabrication. Materials 2020, 13, 2278. [Google Scholar] [CrossRef]
- Oyama, T.G.; Oyama, K.; Kimura, A.; Yoshida, F.; Ishida, R.; Yamazaki, M.; Miyoshi, H.; Taguchi, M. Collagen Hydrogels with Controllable Combined Cues of Elasticity and Topography to Regulate Cellular Processes. Biomed. Mater. 2021, 16, 045037. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Yu, C.; Li, X.; Bao, H.; Zhang, B.; Chen, Z.; Zhang, Z. Facile Engineering of ECM-Mimetic Injectable Dual Crosslinking Hydrogels with Excellent Mechanical Resilience, Tissue Adhesion, and Biocompatibility. J. Mater. Chem. B 2021, 9, 10003–10014. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Zhang, Y.; Xie, J.; Wang, X.; Cao, L.; Chen, G.; Mao, H.; Bi, X.; Gu, Z.; Yang, J. VE-Cadherin Functionalized Injectable PAMAM/HA Hydrogel Promotes Endothelial Differentiation of hMSCs and Vascularization. Appl. Mater. Today 2020, 20, 100690. [Google Scholar] [CrossRef]
- Aldana, A.A.; Valente, F.; Dilley, R.; Doyle, B. Development of 3D Bioprinted GelMA-Alginate Hydrogels with Tunable Mechanical Properties. Bioprinting 2021, 21, e00105. [Google Scholar] [CrossRef]
- Tang, Q.; Lu, B.; He, J.; Chen, X.; Fu, Q.; Han, H.; Luo, C.; Yin, H.; Qin, Z.; Lyu, D.; et al. Exosomes-Loaded Thermosensitive Hydrogels for Corneal Epithelium and Stroma Regeneration. Biomaterials 2022, 280, 121320. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Molino, B.Z.; Wang, X.; Cheng, F.; Xu, W.; Molino, P.; Bacher, M.; Su, D.; Rosenau, T.; Willför, S.; et al. 3D Printing of Nanocellulose Hydrogel Scaffolds with Tunable Mechanical Strength Towards Wound Healing Application. J. Mater. Chem. B 2018, 6, 7066–7075. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Han, S.H.; Kook, Y.-M.; Lee, K.-M.; Jin, Y.-Z.; Koh, W.-G.; Lee, J.H.; Lee, K. A Novel 3D Indirect Co-culture System Based on a Collagen Hydrogel Scaffold for Enhancing the Osteogenesis of Stem Cells. J. Mater. Chem. B 2020, 8, 9481–9491. [Google Scholar] [CrossRef]
- Lu, M.; Liu, Y.; Huang, Y.-C.; Huang, C.-J.; Tsai, W.-B. Fabrication of Photo-Crosslinkable Glycol Chitosan Hydrogel as a Tissue Adhesive. Carbohydr. Polym. 2018, 181, 668–674. [Google Scholar] [CrossRef]
- Shen, Y.; Tang, H.; Huang, X.; Hang, R.; Zhang, X.; Wang, Y.; Yao, X. DLP Printing Photocurable Chitosan to Build Bio-Constructs for Tissue Engineering. Carbohydr. Polym. 2020, 235, 115970. [Google Scholar] [CrossRef]
- Aranaz, I.; Alcántara, A.R.; Civera, M.C.; Arias, C.; Elorza, B.; Caballero, A.H.; Acosta, N. Chitosan: An Overview of Its Properties and Applications. Polymers 2021, 13, 3256. [Google Scholar] [CrossRef]
- Fan, B.; Cui, N.; Xu, Z.; Chen, K.; Yin, P.; Yue, K.; Tang, W. Thermoresponsive and Self-Healing Hydrogel Based on Chitosan Derivatives and Polyoxometalate as an Antibacterial Coating. Biomacromolecules 2022, 23, 972–982. [Google Scholar] [CrossRef] [PubMed]
- Lai, W.-F. Non-conjugated Polymers with Intrinsic Luminescence for Drug Delivery. J. Drug Deliv. Sci. Technol. 2020, 59, 101916. [Google Scholar] [CrossRef]
- Chen, L.; Li, Z.; Zheng, Y.; Zhou, F.; Zhao, J.; Zhai, Q.; Zhang, Z.; Liu, T.; Chen, Y.; Qi, S. 3D-Printed Dermis-Specific Extracellular Matrix Mitigates Scar Contraction via Inducing Early Angiogenesis and Macrophage M2 Polarization. Bioact. Mater. 2022, 10, 236–246. [Google Scholar] [CrossRef] [PubMed]
- Olsen, D.; Yang, C.; Bodo, M.; Chang, R.; Leigh, S.; Baez, J.; Carmichael, D.; Perälä, M.; Hämäläinen, E.-H.; Jarvinen, M.; et al. Recombinant Collagen and Gelatin for Drug Delivery. Adv. Drug Deliv. Rev. 2003, 55, 1547–1567. [Google Scholar] [CrossRef] [PubMed]
- Gibney, R.; Patterson, J.; Ferraris, E. High-Resolution Bioprinting of Recombinant Human Collagen Type III. Polymers 2021, 13, 2973. [Google Scholar] [CrossRef]
- Deng, A.; Yang, Y.; Du, S.; Yang, X.; Pang, S.; Wang, X.; Yang, S. Preparation of a Recombinant Collagen-Peptide (RHC)-Conjugated Chitosan Thermosensitive Hydrogel for Wound Healing. Mater. Sci. Eng. C 2020, 119, 111555. [Google Scholar] [CrossRef]
- Deng, A.; Yang, Y.; Du, S.; Yang, S. Electrospinning of In Situ Crosslinked Recombinant Human Collagen Peptide/Chitosan Nanofibers for Wound Healing. Biomater. Sci. 2018, 6, 2197–2208. [Google Scholar] [CrossRef]
- Deng, A.; Yang, Y.; Du, S. Tissue Engineering 3D Porous Scaffolds Prepared from Electrospun Recombinant Human Collagen (RHC) Polypeptides/Chitosan Nanofibers. Appl. Sci. 2021, 11, 5096. [Google Scholar] [CrossRef]
- Liu, B.; Lei, Y.; Zhang, J.; Hu, L.; Yang, S. Expression, Purification and Characterization of Recombinant Human Gelatin in Pichia pastoris. Appl. Chem. Eng. 2011, 236–238, 2905–2912. [Google Scholar] [CrossRef]
- Oommen, O.P.; Wang, S.; Kisiel, M.; Sloff, M.; Hilborn, J.; Varghese, O.P. Smart Design of Stable Extracellular Matrix Mimetic Hydrogel: Synthesis, Characterization, and In Vitro and In Vivo Evaluation for Tissue Engineering. Adv. Funct. Mater. 2013, 23, 1273–1280. [Google Scholar] [CrossRef]
- Zhao, C.; Wu, Z.; Chu, H.; Wang, T.; Qiu, S.; Zhou, J.; Zhu, Q.; Liu, X.; Quan, D.; Bai, Y. Thiol-Rich Multifunctional Macromolecular Crosslinker for Gelatin-Norbornene-Based Bioprinting. Biomacromolecules 2021, 22, 2729–2739. [Google Scholar] [CrossRef] [PubMed]
- Fernandes Queiroz, M.; Melo, K.R.; Sabry, D.A.; Sassaki, G.L.; Rocha, H.A. Does the Use of Chitosan Contribute to Oxalate Kidney Stone Formation? Mar. Drugs 2014, 13, 141–158. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Krishnaraj, R.; Desai, T.A. Evaluation of Nanostructured Composite Collagen–Chitosan Matrices for Tissue Engineering. Tissue Eng. 2001, 7, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Kalra, A.; Lowe, A.; Al-Jumaily, A.M. Mechanical Behaviour of Skin: A Review. J. Mater. Sci. Eng. 2016, 5, 1000254. [Google Scholar]
- Raftery, R.M.; Woods, B.; Marques, A.L.P.; Moreira-Silva, J.; Silva, T.H.; Cryan, S.-A.; Reis, R.L.; O’Brien, F.J. Multifunctional Biomaterials from the Sea: Assessing the Effects of Chitosan Incorporation into Collagen Scaffolds on Mechanical and Biological Functionality. Acta Biomater. 2016, 43, 160–169. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Ritchie, A.C.; Everitt, N.M. Recombinant Human Collagen/Chitosan-Based Soft Hydrogels as Biomaterials for Soft Tissue Engineering. Mater. Sci. Eng. C 2021, 121, 111846. [Google Scholar] [CrossRef]
- Malekpour, A.; Chen, X. Printability and Cell Viability in Extrusion-Based Bioprinting from Experimental, Computational, and Machine Learning Views. J. Funct. Biomater. 2022, 13, 40. [Google Scholar] [CrossRef]
- Jia, J.; Richards, D.J.; Pollard, S.; Tan, Y.; Rodriguez, J.; Visconti, R.P.; Trusk, T.C.; Yost, M.J.; Yao, H.; Markwald, R.R.; et al. Engineering Alginate as Bioink for Bioprinting. Acta Biomater. 2014, 10, 4323–4331. [Google Scholar] [CrossRef] [Green Version]
- Murphy, S.V.; Skardal, A.; Atala, A. Evaluation of Hydrogels for Bio-Printing Applications. J. Biomed. Mater. Res. Part A 2012, 101A, 272–284. [Google Scholar] [CrossRef]
- Barros, N.R.; Kim, H.-J.; Gouidie, M.J.; Lee, K.; Bandaru, P.; Banton, E.A.; Sarikhani, E.; Sun, W.; Zhang, S.; Cho, H.-J.; et al. Biofabrication of Endothelial Cell, Dermal Fibroblast, and Multilayered Keratinocyte Layers for Skin Tissue Engineering. Biofabrication 2021, 13, 035030. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Wang, Z.; Xu, Y.; Xia, J.; Xu, Z.; Zhu, S.; Jin, M. Preparation of Chitosan/Recombinant Human Collagen-Based Photo-Responsive Bioinks for 3D Bioprinting. Gels 2022, 8, 314. https://0-doi-org.brum.beds.ac.uk/10.3390/gels8050314
Yang Y, Wang Z, Xu Y, Xia J, Xu Z, Zhu S, Jin M. Preparation of Chitosan/Recombinant Human Collagen-Based Photo-Responsive Bioinks for 3D Bioprinting. Gels. 2022; 8(5):314. https://0-doi-org.brum.beds.ac.uk/10.3390/gels8050314
Chicago/Turabian StyleYang, Yang, Zixun Wang, Yuanyuan Xu, Jingjing Xia, Zhaoxian Xu, Shuai Zhu, and Mingjie Jin. 2022. "Preparation of Chitosan/Recombinant Human Collagen-Based Photo-Responsive Bioinks for 3D Bioprinting" Gels 8, no. 5: 314. https://0-doi-org.brum.beds.ac.uk/10.3390/gels8050314