In Vitro and In Silico Characterization of Curcumin-Loaded Chitosan–PVA Hydrogels: Antimicrobial and Potential Wound Healing Activity
Abstract
:1. Introduction
2. Results and Discussion
2.1. Thickness, Weight Variation, and Folding Endurance
2.2. Moisture Content and Moisture Uptake
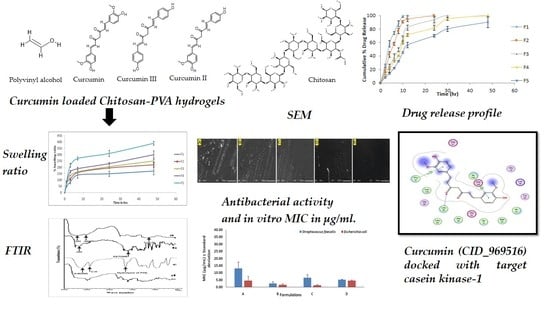
2.3. Swelling Ratio
2.4. WVTR
2.5. Tensile Strength and Elongation at Break
2.6. FTIR (Fourier Transform Infrared) Spectroscopy
2.7. SEM (Scanning Electron Microscopy)
2.8. Antimicrobial Study
2.9. In Vitro Drug Release
2.10. Protein Targets and Ligand Preparations for Analysis
2.11. Molecular Docking
3. Conclusions
4. Materials and Methods
4.1. Materials
4.2. Methods
4.2.1. Thickness and Weight Variation
4.2.2. Folding Endurance
4.2.3. Moisture Content
4.2.4. Moisture Uptake
4.2.5. Swelling Ratio
4.2.6. WVTR
4.2.7. Mechanical Properties
4.2.8. FTIR
4.2.9. SEM
4.2.10. Antimicrobial Activity
4.2.11. In Vitro Drug Release
4.2.12. Selection of Protein Targets and Chemical Compounds
4.2.13. Molecular Docking
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chopra, H.; Gandhi, S.; Gautam, R.K.; Kamal, M.A. Bacterial Nanocellulose Based Wound Dressings: Current and Future Prospects. Curr. Pharm. Des. 2021, 28, 570–580. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.M.; Iqbal, M.; Chopra, H.; Urmi, S.; Junapudi, S.; Bibi, S.; Gupta, S.K.; Pangi, V.N.; Singh, I.; Abdel-Daim, M.M. Pivotal Role of Vitamin D in Mitochondrial Health, Cardiac Function, and Human Reproduction. EXCLI J. 2022, 21, 967–990. [Google Scholar] [CrossRef]
- Chopra, H.; Kumar, S.; Singh, I. Bioadhesive Hydrogels and Their Applications. In Bioadhesives in Drug Delivery; Wiley: Hoboken, NJ, USA, 2020; pp. 147–170. [Google Scholar]
- Chopra, H.; Priyanka; Choudhary, O.P.; Emran, T.B. Three dimensional printed wound dressings: Recent progresses. Int. J. Surg. 2023, 109, 549–550. [Google Scholar] [CrossRef] [PubMed]
- Chopra, H.; Kumar, S.; Singh, I. Strategies and Therapies for Wound Healing: A Review. Curr. Drug Targets 2021, 23, 87–98. [Google Scholar] [CrossRef]
- Singh, I.; Chopra, H.; Kumar, S. Biopolymer Based Scaffolds for Tissue Engineering Applications. Curr. Drug Targets 2020, 21, 282–295. [Google Scholar] [CrossRef]
- Singh Bakshi, I.; Chopra, H.; Sharma, M.; Kaushik, D.; Pahwa, R. Haryanto Herbal Bioactives for Wound Healing Application. In Herbal Bioactive-Based Drug Delivery Systems; Academic Press: Cambridge, MA, USA, 2022; pp. 259–282. [Google Scholar] [CrossRef]
- Dhingra, G.A.; Kaur, M.; Singh, M.; Aggarwal, G.; Nagpal, M. Lock Stock and Barrel of Wound Healing. Curr. Pharm. Des. 2019, 25, 4090–4107. [Google Scholar] [CrossRef]
- Kumar, A.; Behl, T.; Chadha, S. A Rationalized and Innovative Perspective of Nanotechnology and Nanobiotechnology in Chronic Wound Management. J. Drug Deliv. Sci. Technol. 2020, 60, 101930. [Google Scholar] [CrossRef]
- Sharma, P.; Kumar, A.; Agarwal, T.; Dey, A.D.; Moghaddam, F.D.; Rahimmanesh, I.; Ghovvati, M.; Yousefiasl, S.; Borzacchiello, A.; Mohammadi, A.; et al. Nucleic Acid-Based Therapeutics for Dermal Wound Healing. Int. J. Biol. Macromol. 2022, 220, 920–933. [Google Scholar] [CrossRef]
- Elsadek, N.E.; Nagah, A.; Ibrahim, T.M.; Chopra, H.; Ghonaim, G.A.; Emam, S.E.; Cavalu, S.; Attia, M.S. Electrospun Nanofibers Revisited: An Update on the Emerging Applications in Nanomedicine. Materials 2022, 15, 1934. [Google Scholar] [CrossRef]
- Chopra, H.; Kumar, S.; Safi, S.Z.; Singh, I.; Emran, T. Bin Wound Dressings: Recent Updates. Int. J. Surg. 2022, 104, 106793. [Google Scholar] [CrossRef]
- Chopra, H.; Kumar, S.; Singh, I. Bioinks for 3D Printing of Artificial Extracellular Matrices. In Advanced 3D-Printed Systems and Nanosystems for Drug Delivery and Tissue Engineering; Elsevier: Amsterdam, The Netherlands, 2020; pp. 1–37. [Google Scholar] [CrossRef]
- Liang, Y.; He, J.; Guo, B. Functional Hydrogels as Wound Dressing to Enhance Wound Healing. ACS Nano 2021, 15, 12687–12722. [Google Scholar] [CrossRef]
- Qu, J.; Zhao, X.; Liang, Y.; Zhang, T.; Ma, P.X.; Guo, B. Antibacterial Adhesive Injectable Hydrogels with Rapid Self-Healing, Extensibility and Compressibility as Wound Dressing for Joints Skin Wound Healing. Biomaterials 2018, 183, 185–199. [Google Scholar] [CrossRef]
- Yu, R.; Yang, Y.; He, J.; Li, M.; Guo, B. Novel Supramolecular Self-Healing Silk Fibroin-Based Hydrogel via Host–Guest Interaction as Wound Dressing to Enhance Wound Healing. Chem. Eng. J. 2021, 417, 128278. [Google Scholar] [CrossRef]
- Chopra, H.; Dey, P.S.; Das, D.; Bhattacharya, T.; Shah, M.; Mubin, S.; Maishu, S.P.; Akter, R.; Rahman, M.H.; Karthika, C.; et al. Curcumin Nanoparticles as Promising Therapeutic Agents for Drug Targets. Molecules 2021, 26, 4998. [Google Scholar] [CrossRef]
- Jantarat, C. Bioavailability Enhancement Techniques of Herbal Medicine: A Case Example of Curcumin. Int. J. Pharm. Pharm. Sci. 2013, 5, 493–500. [Google Scholar]
- Joe, B.; Vijaykumar, M.; Lokesh, B.R. Biological Properties of Curcumin-Cellular and Molecular Mechanisms of Action. Crit. Rev. Food Sci. Nutr. 2004, 44, 97–111. [Google Scholar] [CrossRef]
- Frey, R.S.; Malik, A.B. Oxidant Signaling in Lung Cells. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2004, 286, L1–L3. [Google Scholar] [CrossRef]
- Gadekar, R.; Saurabh, M.K.; Thakur, G.S.; Saurabh, A. Study of Formulation, Characterisation and Wound Healing Potential of Transdermal Patches of Curcumin. Asian J. Pharm. Clin. Res. 2012, 5, 225–230. [Google Scholar]
- Phan, T.T.; See, P.; Lee, S.T.; Chan, S.Y. Protective Effects of Curcumin against Oxidative Damage on Skin Cells in Vitro: Its Implication for Wound Healing. J. Trauma 2001, 51, 927–931. [Google Scholar] [CrossRef]
- Asai, A.; Miyazawa, T. Occurrence of Orally Administered Curcuminoid as Glucuronide and Glucuronide/Sulfate Conjugates in Rat Plasma. Life Sci. 2000, 67, 2785–2793. [Google Scholar] [CrossRef]
- Anand, P.; Kunnumakkara, A.B.; Newman, R.A.; Aggarwal, B.B. Bioavailability of Curcumin: Problems and Promises. Mol. Pharm. 2007, 4, 807–818. [Google Scholar] [CrossRef] [PubMed]
- Sidhu, G.S.; Singh, A.K.; Thaloor, D.; Banaudha, K.K.; Patnaik, G.K.; Srimal, R.C.; Maheshwari, R.K. Enhancement of Wound Healing by Curcumin in Animals. Wound Repair Regen. 1998, 6, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Sidhu, G.S.; Mani, H.; Gaddipati, J.P.; Singh, A.K.; Seth, P.; Banaudha, K.K.; Patnaik, G.K.; Maheshwari, R.K. Curcumin Enhances Wound Healing in Streptozotocin Induced Diabetic Rats and Genetically Diabetic Mice. Wound Repair Regen. 1999, 7, 362–374. [Google Scholar] [CrossRef] [PubMed]
- Hosseinnejad, M.; Jafari, S.M. Evaluation of Different Factors Affecting Antimicrobial Properties of Chitosan. Int. J. Biol. Macromol. 2016, 85, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Rahman, R.M.; Hrdina, R.; Abdel-Mohsen, A.M.; Fouda, M.M.G.; Soliman, A.Y.; Mohamed, F.K.; Mohsin, K.; Pinto, T.D. Chitin and Chitosan from Brazilian Atlantic Coast: Isolation, Characterization and Antibacterial Activity. Int. J. Biol. Macromol. 2015, 80, 107–120. [Google Scholar] [CrossRef]
- Szymańska, E.; Winnicka, K. Stability of Chitosan—A Challenge for Pharmaceutical and Biomedical Applications. Mar. Drugs 2015, 13, 1819–1846. [Google Scholar] [CrossRef]
- Wang, Q.Z.; Chen, X.G.; Liu, N.; Wang, S.X.; Liu, C.S.; Meng, X.H.; Liu, C.G. Protonation Constants of Chitosan with Different Molecular Weight and Degree of Deacetylation. Carbohydr. Polym. 2006, 65, 194–201. [Google Scholar] [CrossRef]
- Liu, H.; Du, Y.; Wang, X.; Sun, L. Chitosan Kills Bacteria through Cell Membrane Damage. Int. J. Food Microbiol. 2004, 95, 147–155. [Google Scholar] [CrossRef]
- Daza Agudelo, J.I.; Ramirez, M.R.; Henquin, E.R.; Rintoul, I. Modelling of Swelling of PVA Hydrogels Considering Non-Ideal Mixing Behaviour of PVA and Water. J. Mater. Chem. B 2019, 7, 4049–4054. [Google Scholar] [CrossRef]
- Ma, S.; Wang, S.; Li, Q.; Leng, Y.; Wang, L.; Hu, G.H. A Novel Method for Preparing Poly(Vinyl Alcohol) Hydrogels: Preparation, Characterization, and Application. Ind. Eng. Chem. Res. 2017, 56, 7971–7976. [Google Scholar] [CrossRef]
- Cazón, P.; Velázquez, G.; Vázquez, M. Characterization of Bacterial Cellulose Films Combined with Chitosan and Polyvinyl Alcohol: Evaluation of Mechanical and Barrier Properties. Carbohydr. Polym. 2019, 216, 72–85. [Google Scholar] [CrossRef]
- Abdeen, Z. Swelling and Reswelling Characteristics of Cross-Linked Poly(Vinyl Alcohol)/Chitosan Hydrogel Film. J. Dispers. Sci. Technol. 2011, 32, 1337–1344. [Google Scholar] [CrossRef]
- Casey, L.S.; Wilson, L.D. Investigation of Chitosan-PVA Composite Films and Their Adsorption Properties. In Proceedings of the International Conference on Environmental Pollution and Public Health, EPPH 2015, Beijing, China, 10–12 April 2015. [Google Scholar]
- Kanatt, S.R.; Rao, M.S.; Chawla, S.P.; Sharma, A. Active Chitosan-Polyvinyl Alcohol Films with Natural Extracts. Food Hydrocoll. 2012, 29, 290–297. [Google Scholar] [CrossRef]
- Pelissari, F.M.; Grossmann, M.V.E.; Yamashita, F.; Pined, E.A.G. Antimicrobial, Mechanical, and Barrier Properties of Cassava Starch-Chitosan Films Incorporated with Oregano Essential Oil. J. Agric. Food Chem. 2009, 57, 7499–7504. [Google Scholar] [CrossRef]
- Nuutila, K.; Eriksson, E. Moist Wound Healing with Commonly Available Dressings. Adv. Wound Care 2021, 10, 685–698. [Google Scholar] [CrossRef]
- Yang, W.; Fortunati, E.; Bertoglio, F.; Owczarek, J.S.; Bruni, G.; Kozanecki, M.; Kenny, J.M.; Torre, L.; Visai, L.; Puglia, D. Polyvinyl Alcohol/Chitosan Hydrogels with Enhanced Antioxidant and Antibacterial Properties Induced by Lignin Nanoparticles. Carbohydr. Polym. 2018, 181, 275–284. [Google Scholar] [CrossRef]
- Kouchak, M.; Ameri, A.; Naseri, B.; Kargar Boldaji, S. Chitosan and Polyvinyl Alcohol Composite Films Containing Nitrofurazone: Preparation and Evaluation. Iran. J. Basic Med. Sci. 2014, 17, 14–20. [Google Scholar] [CrossRef]
- Koosha, M.; Aalipour, H.; Shirazi, M.J.S.; Jebali, A.; Chi, H.; Hamedi, S.; Wang, N.; Li, T.; Moravvej, H. Physically Crosslinked Chitosan/PVA Hydrogels Containing Honey and Allantoin with Long-Term Biocompatibility for Skin Wound Repair: An In Vitro and In Vivo Study. J. Funct. Biomater. 2021, 12, 61. [Google Scholar] [CrossRef]
- Long, A.M.; Zhao, H.; Huang, X. Structural Basis for the Potent and Selective Inhibition of Casein Kinase 1 Epsilon. J. Med. Chem. 2012, 55, 10307–10311. [Google Scholar] [CrossRef]
- Wagner, F.F.; Bishop, J.A.; Gale, J.P.; Shi, X.; Walk, M.; Ketterman, J.; Patnaik, D.; Barker, D.; Walpita, D.; Campbell, A.J.; et al. Inhibitors of Glycogen Synthase Kinase 3 with Exquisite Kinome-Wide Selectivity and Their Functional Effects. ACS Chem. Biol. 2016, 11, 1952–1963. [Google Scholar] [CrossRef]
- Reshad, R.A.I.; Alam, S.; Raihan, H.B.; Meem, K.N.; Rahman, F.; Zahid, F.; Rafid, M.I.; Rahman, S.M.O.; Omit, S.; Ali, M.H. In Silico Investigations on Curcuminoids from Curcuma Longa as Positive Regulators of the Wnt/β-Catenin Signaling Pathway in Wound Healing. Egypt. J. Med. Hum. Genet. 2021, 22, 65. [Google Scholar] [CrossRef]
- Xiong, Y.; Zhou, L.; Su, Z.; Song, J.; Sun, Q.; Liu, S.S.; Xia, Y.; Wang, Z.; Lu, D. Longdaysin Inhibits Wnt/β-Catenin Signaling and Exhibits Antitumor Activity against Breast Cancer. Onco. Targets. Ther. 2019, 12, 993. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; He, X. Wnt/β-Catenin Signaling: New (and Old) Players and New Insights. Curr. Opin. Cell Biol. 2008, 20, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Cruciat, C.M. Casein Kinase 1 and Wnt/β-Catenin Signaling. Curr. Opin. Cell Biol. 2014, 31, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Nie, X.; Shi, X.; Zhao, J.; Chen, Y.; Yao, Q.; Sun, C.; Yang, J. Regulatory Mechanisms of the Wnt/β-Catenin Pathway in Diabetic Cutaneous Ulcers. Front. Pharmacol. 2018, 9, 1114. [Google Scholar] [CrossRef]
- Bastakoty, D.; Young, P.P. Wnt/β-Catenin Pathway in Tissue Injury: Roles in Pathology and Therapeutic Opportunities for Regeneration. FASEB J. 2016, 30, 3271. [Google Scholar] [CrossRef]
- El-Kased, R.F.; Amer, R.I.; Attia, D.; Elmazar, M.M. Honey-Based Hydrogel: In Vitro and Comparative in Vivo Evaluation for Burn Wound Healing. Sci. Rep. 2017, 7, 9692. [Google Scholar] [CrossRef]
- Vilar, S.; Cozza, G.; Moro, S. Medicinal Chemistry and the Molecular Operating Environment (MOE): Application of QSAR and Molecular Docking to Drug Discovery. Curr. Top. Med. Chem. 2008, 8, 1555–1572. [Google Scholar] [CrossRef]
- Standard Test Method for Determining Water Vapor Transmission Rates through Nonwoven and Plastic Barriers. Available online: https://www.astm.org/d6701-21.html (accessed on 3 December 2022).
- Patel, J.B.; Eliopoulos, G.M.; Jenkins, S.G.; James Lewis, F.S.; Brandi Limbago, P.; Nicolau, D.P.; Robin Patel, F.; Powell, M.; Sandra Richter, F.S.; Jana Swenson, D.M.; et al. Performance Standards for Antimicrobial Susceptibility Testing Performance Standards for Antimicrobial Susceptibility Testing Suggested Citation, 33rd ed.; Clinic and laboratory standards Institute: Wayne, PA, USA, 2016; pp. 100–125. [Google Scholar]
- Bibi, S.; Sakata, K. Current Status of Computer-Aided Drug Design for Type 2 Diabetes. Curr. Comput. Aided. Drug Des. 2016, 12, 167–177. [Google Scholar] [CrossRef]
- Bibi, S.; Hasan, M.M.; Wang, Y.-B.; Papadakos, S.P.; Yu, H. Cordycepin as a Promising Inhibitor of SARS-CoV-2 RNA Dependent RNA Polymerase (RdRp). Curr. Med. Chem. 2021, 29, 152–162. [Google Scholar] [CrossRef]
- Bibi, S.; Sakata, K. An Integrated Computational Approach for Plant-Based Protein Tyrosine Phosphatase Non-Receptor Type 1 Inhibitors. Curr. Comput. Aided. Drug Des. 2017, 13, 319–335. [Google Scholar] [CrossRef]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A.; et al. PubChem Substance and Compound Databases. Nucleic Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef]
- Mendelsohn, L.D. ChemDraw 8 Ultra, Windows and Macintosh Versions. J. Chem. Inf. Comput. Sci. 2004, 44, 2225–2226. [Google Scholar] [CrossRef]
- Chopra, H.; Bibi, S.; Kumar, S.; Khan, M.S.; Kumar, P.; Singh, I. Preparation and Evaluation of Chitosan/PVA Based Hydrogel Films Loaded with Honey for Wound Healing Application. Gels 2022, 8, 111. [Google Scholar] [CrossRef]
- Saleem, U.; Bibi, S.; Shah, M.A.; Ahmad, B.; Saleem, A.; Chauhdary, Z.; Anwar, F.; Javaid, N.; Hira, S.; Akhtar, M.F.; et al. Anti-Parkinson’s Evaluation of Brassica Juncea Leaf Extract and Underlying Mechanism of Its Phytochemicals. Front. Biosci. 2021, 26, 1031–1051. [Google Scholar]
- Yousafi, Q.; Batool, J.; Khan, M.S.; Perveen, T.; Sajid, M.W.; Hussain, A.; Mehmood, A.; Saleem, S. In Silico Evaluation of Food Derived Bioactive Peptides as Inhibitors of Angiotensin Converting Enzyme (ACE). Int. J. Pept. Res. Ther. 2021, 27, 341–349. [Google Scholar] [CrossRef]
- Khan, M.; Patujo, J.; Mushtaq, I.; Ishtiaq, A.; Tahir, M.N.; Bibi, S.; Khan, M.S.; Mustafa, G.; Mirza, B.; Badshah, A.; et al. Anti-Diabetic Potential, Crystal Structure, Molecular Docking, DFT, and Optical-Electrochemical Studies of New Dimethyl and Diethyl Carbamoyl-N, N’-Disubstituted Based Thioureas. J. Mol. Struct. 2021, 1253, 132207. [Google Scholar] [CrossRef]
- Saleem, U.; Shehzad, A.; Shah, S.; Raza, Z.; Shah, M.A.; Bibi, S.; Chauhdary, Z.; Ahmad, B. Antiparkinsonian Activity of Cucurbita Pepo Seeds along with Possible Underlying Mechanism. Metab. Brain Dis. 2021, 36, 1231–1251. [Google Scholar] [CrossRef]













| Formulation Code | Thickness (mm) | Weight Variation (g) | Folding Endurance | Moisture Content (%) | Moisture Uptake (%) |
|---|---|---|---|---|---|
| F1 | 0.039 ± 0.004 | 0.417 ± 0.03 | 345 ± 12 | 17.14 ± 1.12 | 10.28 ± 0.04 |
| F2 | 0.041 ± 0.005 | 0.421 ± 0.05 | 414 ± 14 | 19.14 ± 2.25 | 12.14 ± 1.14 |
| F3 | 0.049 ± 0.008 | 0.454 ± 0.04 | 434 ± 10 | 21.24 ± 1.24 | 13.24 ± 0.15 |
| F4 | 0.054 ± 0.007 | 0.472 ± 0.03 | 433 ± 12 | 22.74 ± 1.14 | 13.75 ± 0.14 |
| F5 | 0.057 ± 0.006 | 0.495 ± 0.02 | 478 ± 14 | 26.72 ± 2.17 | 15.47 ± 0.08 |
| Formulation Code | WVTR (g/m2/Day) | Tensile Strength (N) | Elongation at Break (mm) |
|---|---|---|---|
| F1 | 2710.54 ± 15.96 | 4.14 ± 0.24. | 30.14 ± 1.49 |
| F2 | 2541.12 ± 58.47 | 4.74 ± 0.83 | 32.48 ± 5.14 |
| F3 | 2198.65 ± 65.25 | 21.52 ± 2.84 | 33.65 ± 4.58 |
| F4 | 1895.62 ± 54.51 | 28.65 ± 2.98 | 34.21 ± 2.98 |
| F5 | 1630.70 ± 25.68 | 38.87 ± 5.24 | 35.78 ± 2.58 |
| Sr No. | Formulations | MIC (µg/mL) ± Standard Deviation | |
|---|---|---|---|
| Streptococcus faecalis (Gram Positive) | Escherichia coli (Gram Negative) | ||
| 1 | Curcumin (A) | 13.02 ± 4.51 | 4.55 ± 2.98 |
| 2 | Chitosan + PVA + Curcumin (B) | 2.60 ± 1.13 | 1.30 ± 0.56 |
| 3 | Chitosan + PVA (C) | 6.51 ± 2.26 | 1.62 ± 0.57 |
| 4 | Chloramphenicol (D) | 5.25 ± 0.21 | 4.51 ± 0.46 |
| Ligand Names | PubChem ID | Ligand Structures | Selected Target Protein | |
|---|---|---|---|---|
| CSK- Binding Energies (Kcal/mol) | GSK3B- Binding Energies (Kcal/mol) | |||
| Polyvinyl alcohol | 11199 |  | −3.0805 | −2.6834 |
| Curcumin | 969516 |  | −6.9875 | −6.5164 |
| Curcumin III | 5315472 |  | −6.0156 | −5.9395 |
| Curcumin II | 5469424 |  | −6.6456 | −5.9991 |
| Chitosan | 71853 |  | −10.3979 | −10.5200 |
| Ligand Name | Binding Energy (Kcal/mol) | Binding Interaction | |||
|---|---|---|---|---|---|
| Interacting Residues | Interaction Type | Bond Distance | Bond Energy (Kcal/mol) | ||
| Protein target casein kinase-1 (CK1) [PDB ID: 3UZP] | |||||
| Curcumin (CID_969516) | −6.9875 | O5–N LEU85 (A) 6-ring CA PRO87 (A) 6-ring CD1 ILE 148(A) | H-acceptor Pi-H Pi-H | 3.22 4.01 4.44 | −0.9 −0.6 −0.5 |
| Chitosan (CID_71853) | −10.3979 | N48–OD2 ASP149 (A) C102–SD MET82 (A) O34–N GLY215 (A) O35–NZ LYS38 (A) | H-donor H-donor H-acceptor H-acceptor | 3.18 3.90 3.17 3.40 | −0.5 −0.5 −1.2 −0.6 |
| Protein target glycogen synthase kinase-3β (GSK3B) [PDB ID: 5HLP] | |||||
| Curcumin (CID_969516) | −6.5164 | O3–OVAL135 (A) | H-donor | 3.16 | −1.0 |
| Chitosan (CID_71853) | −10.5200 | O3–NZ LYS183 (A) O22–OH TYR134 (A) O25–OG SER66 (A) | H-acceptor H-acceptor H-acceptor | 3.55 3.00 2.93 | −0.6 −1.4 −1.4 |
| Formulation Code | Chitosan Solution | PVA Solution (5% w/v) (mL) | Curcumin (mg) | |
|---|---|---|---|---|
| Concentration of Chitosan (% w/v) | Amount of Chitosan (mL) | |||
| F1 | 0.25 | 80 | 20 | 100 |
| F2 | 0.50 | 80 | 20 | 100 |
| F3 | 0.75 | 80 | 20 | 100 |
| F4 | 1.0 | 80 | 20 | 100 |
| F5 | 2.0 | 80 | 20 | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chopra, H.; Bibi, S.; Mohanta, Y.K.; Kumar Mohanta, T.; Kumar, S.; Singh, I.; Saad Khan, M.; Ranjan Rauta, P.; Alshammari, A.; Alharbi, M.; et al. In Vitro and In Silico Characterization of Curcumin-Loaded Chitosan–PVA Hydrogels: Antimicrobial and Potential Wound Healing Activity. Gels 2023, 9, 394. https://0-doi-org.brum.beds.ac.uk/10.3390/gels9050394
Chopra H, Bibi S, Mohanta YK, Kumar Mohanta T, Kumar S, Singh I, Saad Khan M, Ranjan Rauta P, Alshammari A, Alharbi M, et al. In Vitro and In Silico Characterization of Curcumin-Loaded Chitosan–PVA Hydrogels: Antimicrobial and Potential Wound Healing Activity. Gels. 2023; 9(5):394. https://0-doi-org.brum.beds.ac.uk/10.3390/gels9050394
Chicago/Turabian StyleChopra, Hitesh, Shabana Bibi, Yugal Kishore Mohanta, Tapan Kumar Mohanta, Sandeep Kumar, Inderbir Singh, Muhammad Saad Khan, Pradipta Ranjan Rauta, Abdulrahman Alshammari, Metab Alharbi, and et al. 2023. "In Vitro and In Silico Characterization of Curcumin-Loaded Chitosan–PVA Hydrogels: Antimicrobial and Potential Wound Healing Activity" Gels 9, no. 5: 394. https://0-doi-org.brum.beds.ac.uk/10.3390/gels9050394