Molecular Insights into the Enhanced Activity and/or Thermostability of PET Hydrolase by D186 Mutations
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification of Key Non-Active Site Residue
2.2. Catalytic Activity of D186 Variants
2.3. Thermostability of D186 Variants
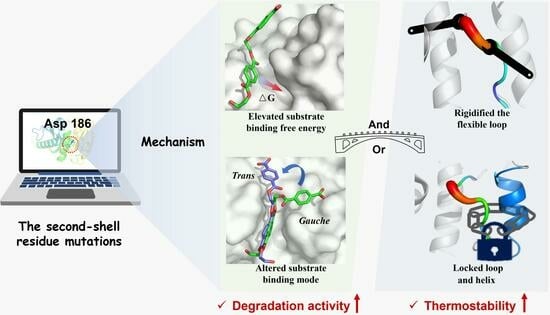
2.4. Molecular Mechanism of Enhanced Catalytic Activity by D186 Mutations
2.5. Molecular Mechanism of Enhanced Thermostability by D186 Mutations
3. Materials and Methods
3.1. General Information
3.2. Site-Directed Mutagenesis, Protein Expression, and Purification
3.3. Enzyme Activity Assay for BHET and PET Film Degradation
3.4. Assay of Enzyme Thermostability
3.5. CD and Fluorescence Spectroscopy
3.6. Molecular Docking and Molecular Dynamics (MD) Simulations
3.7. Measurement of PET Crystallinity
3.8. HPLC Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Moog, D.; Schmitt, J.; Senger, J.; Zarzycki, J.; Rexer, K.H.; Linne, U.; Erb, T.J.; Maier, U.G. Using a marine microalga as a chassis for polyethylene terephthalate (PET) degradation. Microb. Cell Fact. 2020, 18, 171. [Google Scholar] [CrossRef] [PubMed]
- Sagong, H.Y.; Seo, H.; Kim, T.; Son, H.F.; Joo, S.; Lee, S.H.; Kim, S.; Woo, J.S.; Hwang, S.Y.; Kim, K.J. Decomposition of the PET Film by MHETase Using Exo-PETase Function. ACS Catal. 2020, 10, 4805–4812. [Google Scholar] [CrossRef]
- Kumar, A.G.; Anjana, K.; Hinduja, M.; Sujitha, K.; Dharani, G. Review on plastic wastes in marine environment-Biodegradation and biotechnological solutions. Mar. Pollut. Bull. 2020, 150, 110733. [Google Scholar] [CrossRef]
- Joo, S.; Cho, I.J.; Seo, H.; Son, H.F.; Sagong, H.Y.; Shin, T.J.; Choi, S.Y.; Lee, S.Y.; Kim, K.J. Structural insight into molecular mechanism of poly(ethylene terephthalate) degradation. Nat. Commun. 2018, 9, 382. [Google Scholar] [CrossRef]
- Geyer, R.; Jambeck, J.R.; Law, K.L. Production, use, and fate of all plastics ever made. Sci. Adv. 2017, 3, e1700782. [Google Scholar] [CrossRef]
- Gibb, B.C. Plastics are forever. Nat. Chem. 2019, 11, 394–395. [Google Scholar] [CrossRef]
- Magalhaes, R.P.; Fernandes, H.S.; Sousa, S.F. The critical role of Asp206 stabilizing residues on the catalytic mechanism of the Ideonella sakaiensis PETase. Catal. Sci. Technol. 2022, 12, 3474–3483. [Google Scholar] [CrossRef]
- Webb, H.K.; Arnott, J.; Crawford, R.J.; Ivanova, E.P. Plastic Degradation and Its Environmental Implications with Special Reference to Poly(ethylene terephthalate). Polymers 2013, 5, 1–18. [Google Scholar] [CrossRef]
- Gong, J.X.; Kong, T.T.; Li, Y.Q.; Li, Q.J.; Li, Z.; Zhang, J.F. Biodegradation of Microplastic Derived from Poly(ethylene terephthalate) with Bacterial Whole-Cell Biocatalysts. Polymers 2018, 10, 1326. [Google Scholar] [CrossRef]
- Taniguchi, I.; Yoshida, S.; Hiraga, K.; Miyamoto, K.; Kimura, Y.; Oda, K. Biodegradation of PET: Current Status and Application Aspects. ACS Catal. 2019, 9, 4089–4105. [Google Scholar] [CrossRef]
- Jerves, C.; Neves, R.P.P.; Ramos, M.J.; da Silva, S.; Fernandes, P.A. Reaction Mechanism of the PET Degrading Enzyme PETase Studied with DFT/MM Molecular Dynamics Simulations. ACS Catal. 2021, 11, 11626–11638. [Google Scholar] [CrossRef]
- Danso, D.; Schmeisser, C.; Chow, J.; Zimmermann, W.; Wei, R.; Leggewie, C.; Li, X.Z.; Hazen, T.; Streit, W.R. New Insights into the Function and Global Distribution of Polyethylene Terephthalate (PET)-Degrading Bacteria and Enzymes in Marine and Terrestrial Metagenomes. Appl. Environ. Microbiol. 2018, 84, e02773-17. [Google Scholar] [CrossRef]
- Muller, R.J.; Schrader, H.; Profe, J.; Dresler, K.; Deckwer, W.D. Enzymatic degradation of poly(ethylene terephthalate): Rapid hydrolyse using a hydrolase from T-fusca. Macromol. Rapid Commun. 2005, 26, 1400–1405. [Google Scholar] [CrossRef]
- Liu, B.; He, L.H.; Wang, L.P.; Li, T.; Li, C.C.; Liu, H.Y.; Luo, Y.Z.; Bao, R. Protein Crystallography and Site-Direct Mutagenesis Analysis of the Poly(ethylene terephthalate) Hydrolase PETase from Ideonella sakaiensis. ChemBioChem 2018, 19, 1471–1475. [Google Scholar] [CrossRef]
- Bell, E.L.; Smithson, R.; Kilbride, S.; Foster, J.; Hardy, F.J.; Ramachandran, S.; Tedstone, A.A.; Haigh, S.J.; Garforth, A.A.; Day, P.J.R.; et al. Directed evolution of an efficient and thermostable PET depolymerase. Nat. Catal. 2022, 5, 673–681. [Google Scholar] [CrossRef]
- Yoshida, S.; Hiraga, K.; Takehana, T.; Taniguchi, I.; Yamaji, H.; Maeda, Y.; Toyohara, K.; Miyamoto, K.; Kimura, Y.; Oda, K. A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 2016, 351, 1196–1199. [Google Scholar] [CrossRef]
- Son, H.F.; Cho, I.J.; Joo, S.; Seo, H.; Sagong, H.-Y.; Choi, S.Y.; Lee, S.Y.; Kim, K.-J. Rational Protein Engineering of Thermo-Stable PETase from Ideonella sakaiensis for Highly Efficient PET Degradation. ACS Catal. 2019, 9, 3519–3526. [Google Scholar] [CrossRef]
- Son, H.F.; Joo, S.; Seo, H.; Sagong, H.Y.; Lee, S.H.; Hong, H.; Kim, K.J. Structural bioinformatics-based protein engineering of thermo-stable PETase from Ideonella sakaiensis. Enzym. Microb. Technol. 2020, 141, 109656. [Google Scholar] [CrossRef]
- Feng, S.S.; Yue, Y.; Zheng, M.N.; Li, Y.W.; Zhang, Q.Z.; Wang, W.X. IsPETase- and IsMHETase-Catalyzed Cascade Degradation Mechanism toward Polyethylene Terephthalate. ACS Sustain. Chem. Eng. 2021, 9, 9823–9832. [Google Scholar] [CrossRef]
- Cui, Y.L.; Chen, Y.C.; Liu, X.Y.; Dong, S.J.; Tian, Y.E.; Qiao, Y.X.; Mitra, R.; Han, J.; Li, C.L.; Han, X.; et al. Computational Redesign of a PETase for Plastic Biodegradation under Ambient Condition by the GRAPE Strategy. ACS Catal. 2021, 11, 1340–1350. [Google Scholar] [CrossRef]
- Lu, H.Y.; Diaz, D.J.; Czarnecki, N.J.; Zhu, C.Z.; Kim, W.T.; Shroff, R.; Acosta, D.J.; Alexander, B.R.; Cole, H.O.; Zhang, Y.; et al. Machine learning-aided engineering of hydrolases for PET depolymerization. Nature 2022, 604, 662–667. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.X.; Liu, P.; Tan, Z.J.; Zhao, W.; Gao, J.F.; Gu, Q.; Ma, H.W.; Liu, H.F.; Zhu, L.L. Complete Depolymerization of PET Wastes by an Evolved PET Hydrolase from Directed Evolution. Angew. Chem. Int. Edit 2023, 62, e202218390. [Google Scholar] [CrossRef]
- Zhang, Y.F. A relay for improving the catalytic efficiency and thermostability of PET hydrolases. Chem. Catal. 2022, 2, 2420–2422. [Google Scholar] [CrossRef]
- Anishchenko, I.; Ovchinnikov, S.; Kamisetty, H.; Baker, D. Origins of coevolution between residues distant in protein 3D structures. Proc. Natl. Acad. Sci. USA 2017, 114, 9122–9127. [Google Scholar] [CrossRef]
- Truong, D.P.; Rousseau, S.; Machala, B.W.; Huddleston, J.P.; Zhu, M.Z.; Hull, K.G.; Romo, D.; Raushel, F.M.; Sacchettini, J.C.; Glasner, M.E. Second-Shell Amino Acid R266 Helps Determine N-Succinylamino Acid Racemase Reaction Specificity in Promiscuous N-Succinylamino Acid Racemase/o-Succinylbenzoate Synthase Enzymes. Biochemistry 2021, 60, 3829–3840. [Google Scholar] [CrossRef]
- Lee, J.; Goodey, N.M. Catalytic Contributions from Remote Regions of Enzyme Structure. Chem. Rev. 2011, 111, 7595–7624. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Xu, Y.; Nie, Y. Role of distal sites in enzyme engineering. Biotechnol. Adv. 2023, 63, 108094. [Google Scholar] [CrossRef]
- Bi, J.; Chen, S.; Zhao, X.; Nie, Y.; Xu, Y. Computation-aided engineering of starch-debranching pullulanase from Bacillus thermoleovorans for enhanced thermostability. Appl. Microbiol. Biotechnol. 2020, 104, 7551–7562. [Google Scholar] [CrossRef]
- Parasuram, R.; Coulther, T.A.; Hollander, J.M.; Keston-Smith, E.; Ondrechen, M.J.; Beuning, P.J. Prediction of Active Site and Distal Residues in E. coli DNA Polymerase III alpha Polymerase Activity. Biochemistry 2018, 57, 1063–1072. [Google Scholar] [CrossRef]
- Ma, E.J.; Siirola, E.; Moore, C.; Kummer, A.; Stoeckli, M.; Faller, M.; Bouquet, C.; Eggimann, F.; Ligibel, M.; Huynh, D.; et al. Machine-Directed Evolution of an Imine Reductase for Activity and Stereoselectivity. ACS Catal. 2021, 11, 12433–12445. [Google Scholar] [CrossRef]
- Yin, Q.D.; You, S.P.; Zhang, J.X.; Qi, W.; Su, R.X. Enhancement of the polyethylene terephthalate and mono-(2-hydroxyethyl) terephthalate degradation activity of Ideonella sakaiensis PETase by an electrostatic interaction-based strategy. Bioresour. Technol. 2022, 364, 128026. [Google Scholar] [CrossRef]
- Qu, Z.; Chen, K.; Zhang, L.; Sun, Y. Computation-Based Design of Salt Bridges in PETase for Enhanced Thermostability and Performance for PET Degradation. ChemBioChem 2023, 21, e202300373. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11 Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Wei, R.; Breite, D.; Song, C.; Gräsing, D.; Ploss, T.; Hille, P.; Schwerdtfeger, R.; Matysik, J.; Schulze, A.; Zimmermann, W. Biocatalytic Degradation Efficiency of Postconsumer Polyethylene Terephthalate Packaging Determined by Their Polymer Microstructures. Adv. Sci. 2019, 6, 1900491. [Google Scholar] [CrossRef] [PubMed]
- Espino-Rammer, L.; Ribitsch, D.; Przylucka, A.; Marold, A.; Greimel, K.J.; Acero, E.H.; Guebitz, G.M.; Kubicek, C.P.; Druzhinina, I.S. Two Novel Class II Hydrophobins from spp. Stimulate Enzymatic Hydrolysis of Poly(Ethylene Terephthalate) when Expressed as Fusion Proteins. Appl. Environ. Microbiol. 2013, 79, 4230–4238. [Google Scholar] [CrossRef] [PubMed]
- Inaba, S.; Kamiya, N.; Bekker, G.J.; Kawai, F.; Oda, M. Folding thermodynamics of PET-hydrolyzing enzyme Cut190 depending on Ca concentration. J. Therm. Anal. Calorim. 2019, 135, 2655–2663. [Google Scholar] [CrossRef]
- Bollinger, A.; Thies, S.; Knieps-Grünhagen, E.; Gertzen, C.; Kobus, S.; Höppner, A.; Ferrer, M.; Gohlke, H.; Smits, S.H.J.; Jaeger, K.E. A Novel Polyester Hydrolase from the Marine Bacterium-Structural and Functional Insights. Front. Microb. 2020, 11, 114. [Google Scholar] [CrossRef] [PubMed]
- Meilleur, C.; Hupé, J.F.; Juteau, P.; Shareck, F. Isolation and characterization of a new alkali-thermostable lipase cloned from a metagenomic library. J. Ind. Microbiol. Biotechnol. 2009, 36, 853–861. [Google Scholar] [CrossRef]
- Chen, C.C.; Han, X.; Li, X.; Jiang, P.C.; Niu, D.; Ma, L.X.; Liu, W.D.; Li, S.Y.; Qu, Y.Y.; Hu, H.B.; et al. General features to enhance enzymatic activity of poly(ethylene terephthalate) hydrolysis. Nat. Catal. 2021, 4, 425–430. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Fesko, K.; Suplatov, D.; Svedas, V. Bioinformatic analysis of the fold type I PLP-dependent enzymes reveals determinants of reaction specificity in L-threonine aldolase from. FEBS Open Bio 2018, 8, 1013–1028. [Google Scholar] [CrossRef] [PubMed]
- Suplatov, D.; Shalaeva, D.; Kirilin, E.; Arzhanik, V.; Svedas, V. Bioinformatic analysis of protein families for identification of variable amino acid residues responsible for functional diversity. J. Biomol. Struct. Dyn. 2014, 32, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Suplatov, D.; Voevodin, V.; Svedas, V. Robust enzyme design: Bioinformatic tools for improved protein stability. Biotechnol. J. 2015, 10, 344–355. [Google Scholar] [CrossRef]
- Pleiss, J. Systematic Analysis of Large Enzyme Families: Identification of Specificity- and Selectivity-Determining Hotspots. ChemCatChem 2014, 6, 944–950. [Google Scholar] [CrossRef]
- Chen, K.; Hu, Y.; Dong, X.Y.; Sun, Y. Molecular Insights into the Enhanced Performance of EKylated PETase Toward PET Degradation. ACS Catal. 2021, 11, 7358–7370. [Google Scholar] [CrossRef]
- Han, X.; Liu, W.; Huang, J.W.; Ma, J.; Zheng, Y.; Ko, T.P.; Xu, L.; Cheng, Y.S.; Chen, C.C.; Guo, R.T. Structural insight into catalytic mechanism of PET hydrolase. Nat. Commun. 2017, 8, 2106. [Google Scholar] [CrossRef]
- Eiamthong, B.; Meesawat, P.; Wongsatit, T.; Jitdee, J.; Sangsri, R.; Patchsung, M.; Aphicho, K.; Suraritdechachai, S.; Huguenin-Dezot, N.; Tang, S.; et al. Discovery and Genetic Code Expansion of a Polyethylene Terephthalate (PET) Hydrolase from the Human Saliva Metagenome for the Degradation and Bio-Functionalization of PET. Angew. Chem. Int. Edit 2022, 61, e202203061. [Google Scholar] [CrossRef]
- Fecker, T.; Galaz-Davison, P.; Engelberger, F.; Narui, Y.; Sotomayor, M.; Parra, L.P.; Ramirez-Sarmiento, C.A. Active Site Flexibility as a Hallmark for Efficient PET Degradation by I-sakaiensis PETase. Biophys. J. 2018, 114, 1302–1312. [Google Scholar] [CrossRef]
- Kumari, R.; Kumar, R.; Lynn, A.; Consort, O.S.D.D. g_mmpbsa-A GROMACS Tool for High-Throughput MM-PBSA Calculations. J. Chem. Inf. Model 2014, 54, 1951–1962. [Google Scholar] [CrossRef]
- Chen, K.; Dong, X.Y.; Sun, Y. Sequentially co-immobilized PET and MHET hydrolases via Spy chemistry in calcium phosphate nanocrystals present high-performance PET degradation. J. Hazard. Mater. 2022, 438, 129517. [Google Scholar] [CrossRef]
- Wei, R.; Song, C.; Gräsing, D.; Schneider, T.; Bielytskyi, P.; Böttcher, D.; Matysik, J.; Bornscheuer, U.T.; Zimmermann, W. Conformational fitting of a flexible oligomeric substrate does not explain the enzymatic PET degradation. Nat. Commun. 2019, 10, 5581. [Google Scholar] [CrossRef]
- Guo, B.Y.; Vanga, S.R.; Lopez-Lorenzo, X.; Saenz-Mendez, P.; Ericsson, S.R.; Fang, Y.; Ye, X.C.; Schriever, K.; Backstrom, E.; Biundo, A.; et al. Conformational Selection in Biocatalytic Plastic Degradation by PETase. ACS Catal. 2022, 12, 3397–3409. [Google Scholar] [CrossRef]
- Prasad, A.R.; Luduena, R.F.; Horowitz, P.M. Detection of energy transfer between tryptophan residues in the tubulin molecule and bound bis(8-anilinonaphthalene-1-sulfonate), an inhibitor of microtubule assembly, that binds to a flexible region on tubulin. Biochemistry 1986, 25, 3536–3540. [Google Scholar] [CrossRef]
- Lopez-Chavez, E.; Perez-Hernandez, G.; Aparicio, F.; Alas, S.J. On the Thermal Stability of O-6-Methylguanine-DNA Methyltransferase from Archaeon Pyrococcus kodakaraensis by Molecular Dynamics Simulations. J. Chem. Inf. Model 2020, 60, 2138–2154. [Google Scholar] [CrossRef]
- Baker, E.N.; Hubbard, R.E. Hydrogen bonding in globular proteins. Prog. Biophys. Mol. Biol. 1984, 44, 97–179. [Google Scholar] [CrossRef] [PubMed]
- Pierce, A.C.; Sandretto, K.L.; Bemis, G.W. Kinase inhibitors and the case for CH...O hydrogen bonds in protein-ligand binding. Proteins 2002, 49, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Quan, M.Q.; Dong, X.Y.; Shi, Q.H.; Sun, Y. Low modification of PETase enhances its activity toward degrading PET: Effect of conjugate monomer property. Biochem. Eng. J. 2021, 175, 108151. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, Y.; Sun, Y.; Dong, X. Complicated effects of a zwitterionic polymer containing dimethyl chains on the structures, activities and stabilities of different enzymes. Biochem. Eng. J. 2021, 165, 107813. [Google Scholar] [CrossRef]
- Austin, H.P.; Allen, M.D.; Donohoe, B.S.; Rorrer, N.A.; Kearns, F.L.; Silveira, R.L.; Pollard, B.C.; Dominick, G.; Duman, R.; El Omari, K.; et al. Characterization and engineering of a plastic-degrading aromatic polyesterase. Proc. Natl. Acad. Sci. USA 2018, 115, E4350–E4357. [Google Scholar] [CrossRef] [PubMed]
- Kutzner, C.; Páll, S.; Fechner, M.; Esztermann, A.; de Groot, B.L.; Grubmüller, H. Best bang for your buck: GPU nodes for GROMACS biomolecular simulations. J. Comput. Chem. 2015, 36, 1990–2008. [Google Scholar] [CrossRef]
- Kutzner, C.; Páll, S.; Fechner, M.; Esztermann, A.; de Groot, B.L.; Grubmüller, H. More bang for your buck: Improved use of GPU nodes for GROMACS 2018. J. Comput. Chem. 2019, 40, 2418–2431. [Google Scholar] [CrossRef] [PubMed]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 2010, 78, 1950–1958. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. Software News and Update AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.J.; Zhang, L.; Sun, Y. Rational Design of Disulfide Bridges in BbPETase(CD) for Enhancing the Enzymatic Performance in PET Degradation. Molecules 2023, 28, 3528. [Google Scholar] [CrossRef]
- Meng, X.X.; Yang, L.X.; Liu, H.Q.; Li, Q.B.; Xu, G.S.; Zhang, Y.; Guan, F.F.; Zhang, Y.H.; Zhang, W.; Wu, N.F.; et al. Protein engineering of stable IsPETase for PET plastic degradation by Premuse. Int. J. Biol. Macromol. 2021, 180, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Zhong-Johnson, E.Z.L.; Voigt, C.A.; Sinskey, A.J. An absorbance method for analysis of enzymatic degradation kinetics of poly(ethylene terephthalate) films. Sci. Rep. 2021, 11, 2045–2322. [Google Scholar] [CrossRef]
- Enhancement of the degradation capacity of IsPETase for PET plastic degradation by protein engineering. Sci. Total Environ. 2022, 834, 154947. [CrossRef]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qu, Z.; Zhang, L.; Sun, Y. Molecular Insights into the Enhanced Activity and/or Thermostability of PET Hydrolase by D186 Mutations. Molecules 2024, 29, 1338. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules29061338
Qu Z, Zhang L, Sun Y. Molecular Insights into the Enhanced Activity and/or Thermostability of PET Hydrolase by D186 Mutations. Molecules. 2024; 29(6):1338. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules29061338
Chicago/Turabian StyleQu, Zhi, Lin Zhang, and Yan Sun. 2024. "Molecular Insights into the Enhanced Activity and/or Thermostability of PET Hydrolase by D186 Mutations" Molecules 29, no. 6: 1338. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules29061338