Comparative Studies of 5S rDNA Profiles and Cyt b Sequences in two Onychostoma Species (Cyprinidae)
Abstract
:1. Introduction

2. Results and Discussion
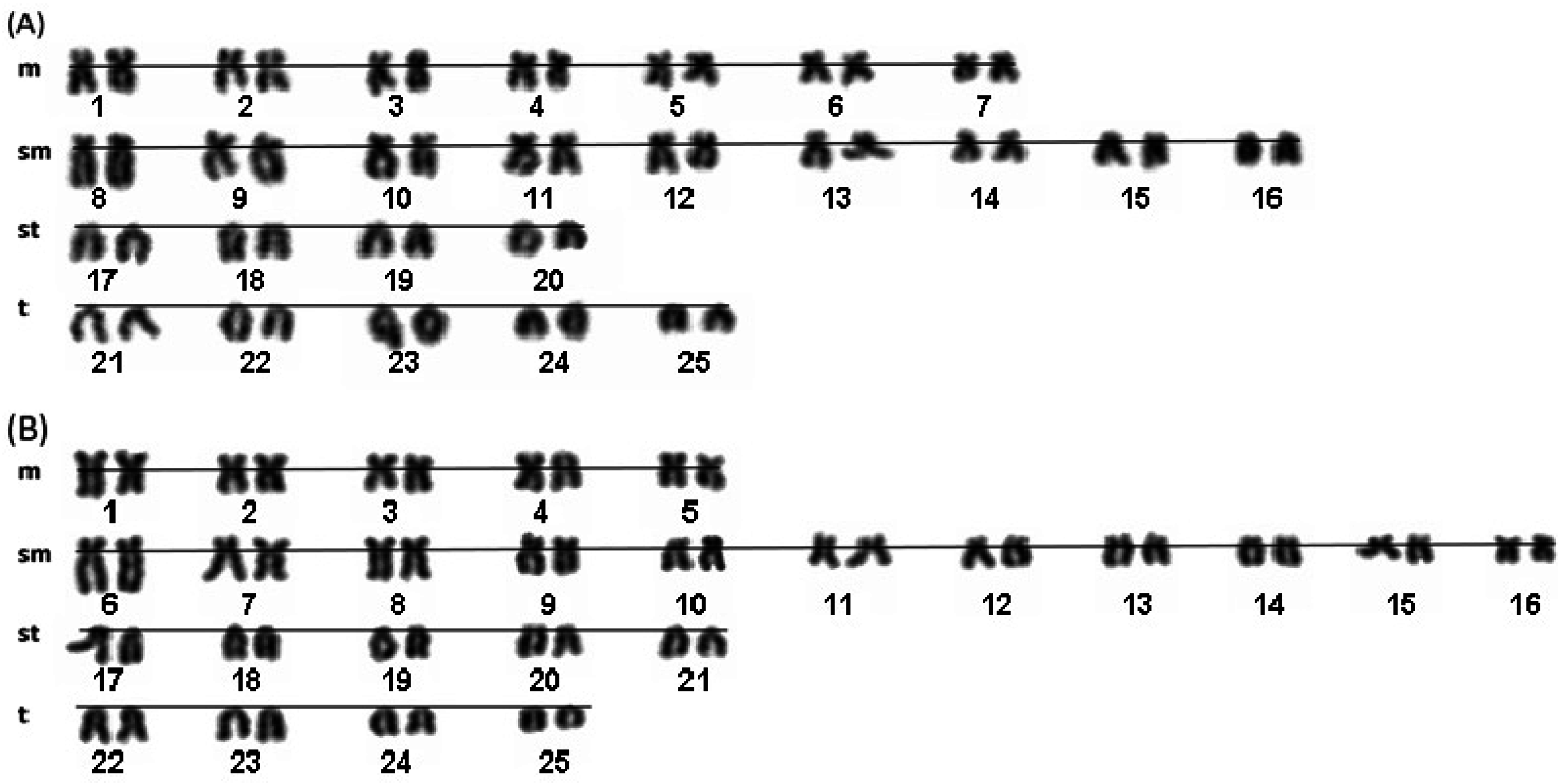


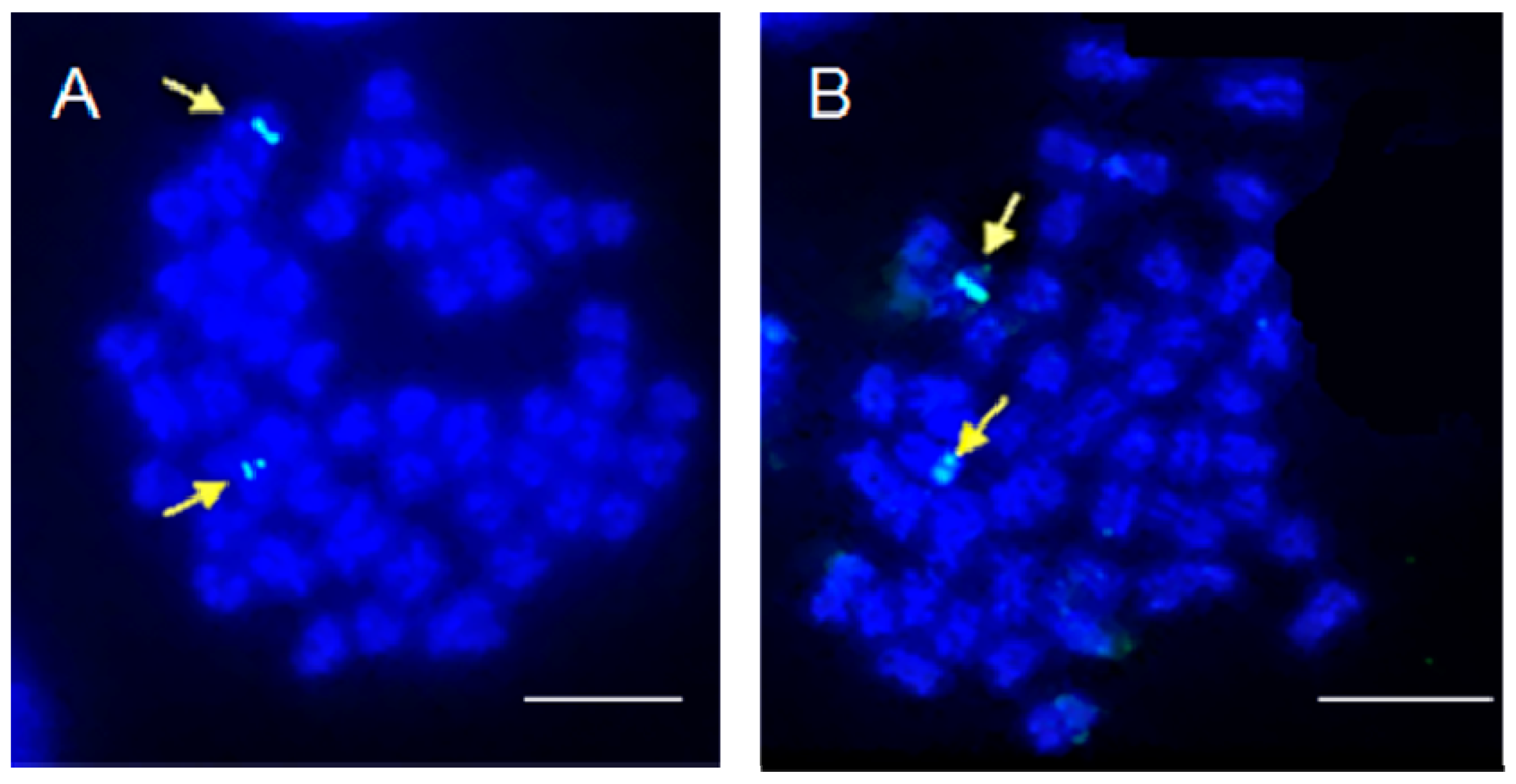

| A1 | A2 and A3 | A4 | A5 | B2 | B1 and B3 | B4 | B5 | Osi | Oov | Vma | Vbe | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | 0.001 | 0.006 | 0.002 | 0.136 | 0.134 | 0.134 | 0.133 | 0.065 | 0.097 | 0.161 | 0.168 | |
| A2 and A3 | 1 | 0.005 | 0.001 | 0.135 | 0.133 | 0.133 | 0.132 | 0.064 | 0.096 | 0.160 | 0.167 | |
| A4 | 7 | 6 | 0.006 | 0.142 | 0.140 | 0.140 | 0.138 | 0.070 | 0.102 | 0.167 | 0.174 | |
| A5 | 2 | 1 | 7 | 0.136 | 0.134 | 0.134 | 0.133 | 0.065 | 0.097 | 0.159 | 0.168 | |
| B2 | 139 | 138 | 144 | 139 | 0.002 | 0.004 | 0.003 | 0.129 | 0.130 | 0.192 | 0.178 | |
| B1 and B3 | 137 | 136 | 142 | 137 | 2 | 0.002 | 0.001 | 0.127 | 0.128 | 0.191 | 0.178 | |
| B4 | 137 | 136 | 142 | 137 | 4 | 2 | 0.001 | 0.127 | 0.128 | 0.191 | 0.179 | |
| B5 | 136 | 135 | 141 | 136 | 3 | 1 | 1 | 0.125 | 0.127 | 0.190 | 0.178 | |
| Osi | 70 | 69 | 75 | 70 | 132 | 130 | 130 | 129 | 0.095 | 0.166 | 0.169 | |
| Oov | 102 | 101 | 107 | 102 | 133 | 131 | 131 | 130 | 100 | 0.164 | 0.172 | |
| Vma | 162 | 161 | 167 | 160 | 188 | 187 | 187 | 186 | 166 | 164 | 0.105 | |
| Vbe | 168 | 167 | 173 | 168 | 177 | 177 | 178 | 177 | 169 | 171 | 109 |
3. Experimental Section
3.1. Fish Collection
3.2. Chromosome Preparations and Staining
3.3. 5S rDNA Purification, Subcloning and Analysis
3.4. Fluorescence in Situ Hybridization of 5S rDNA Probe
3.5. Cytochrome b Gene Cloning and Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wang, J.P.; Lin, H.D.; Huang, S.; Pan, C.H.; Chen, X.L.; Chiang, T.Y. Phylogeography of Varicorhinus barbatulus (Cyprinidae) in Taiwan based on nucleotide variation of mtDNA and allozymes. Mol. Phylogenet. Evol. 2004, 31, 1143–1156. [Google Scholar] [CrossRef] [PubMed]
- Tzeng, C.S. Distribution of the freshwater fishes of Taiwan. J. Taiwan Mus. 1986, 39, 127–146. [Google Scholar]
- Shen, S.C. Fishes of Taiwan; Department of Zoology, National Taiwan University: Taipei, Taiwan, 1993. [Google Scholar]
- Han, C.C.; Tew, K.S.; Chen, I.S.; Su, L.Y.; Fang, L.S. Environmental biology of an endemic cyprinid, Varicorhinus alticorpus, in a subtropical mountain stream of Taiwan. Environ. Biol. Fishes 2000, 59, 153–161. [Google Scholar] [CrossRef]
- Eichler, E.E.; Sankoff, D. Structural dynamics of eukaryotic chromosome evolution. Science 2003, 301, 793–797. [Google Scholar] [CrossRef] [PubMed]
- Foresti de Almeida-Toledo, L.; Daniel-Silva, M.Z.; Moysés, C.B.; Foresti, F. Chromosome variability in Gymnotiformes (Teleostei: Ostariophysi). In Fish Cytogenetics; Pisano, E., Ozouf-Costaz, C., Foresti, F., Kapoor, B.G., Eds.; CRC Press: Boca Raton, FL, USA, 2007; pp. 17–40. [Google Scholar]
- Dai, Y. Karyotype and evolution analysis of vulnerable fish Onychostoma lini from China. In Proceedings of the 7th International Conference on Systems Biology, Huangshan, China, 23–25 August 2013.
- Li, Y.; Li, K.; Jiang, J.; Sun, Q.; Zhou, T. Studies on the karyotypes of Chinese cyprinid fishes X. Karyotypes of five species of barbinae and four species of gobioninae. Zool. Res. 1986, 7, 183–189. [Google Scholar]
- Gui, J.; Li, Y.; Li, K.; Zhou, T. Studies on karyotypes of Chinese cyprinid fishes. VIII. Karyotype analyses of fifteen species of barbines with considerations for their phyletic evolution. Trans. Chin. Ichthyol. Soc. 1986, 5, 119–127. [Google Scholar]
- Phillips, R.B. Application of fluorescence in situ hybridization (FISH) to genome mapping in fishes. In Fish Cytogenetics; Pisano, E., Ozouf-Costaz, C., Foresti, F., Kapoor, B.G., Eds.; CRC Press: Boca Raton, FL, USA, 2007; pp. 455–471. [Google Scholar]
- Martins, C. Chromosomes and Repetitive DNAs: A contribution to the knowledge of the fish genome. In Fish Cytogenetics; Pisano, E., Ozouf-Costaz, C., Foresti, F., Kapoor, B.G., Eds.; CRC Press: Boca Raton, FL, USA, 2007; pp. 421–432. [Google Scholar]
- Wasko, A.P.; Martins, C.; Wright, J.M.; Galetti, P.M., Jr. Molecular organization of 5S rDNA in fishes of the genus Brycon. Genome 2001, 44, 893–902. [Google Scholar] [CrossRef] [PubMed]
- Affonso, P.R.A.M.; Galetti, P.M., Jr. Chromosomal diversification of reef fishes from genus Centropyge (Perciformes, Pomacanthidae). Genetica 2005, 123, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Van Campenhout, S.; Aert, R.; Volckaert, G. Orthologous DNA sequence variation among 5S ribosomal RNA gene spacer sequences on homologous chromosomes 1B, 1D, and 1R of wheat and rye. Genome 1998, 41, 244–255. [Google Scholar] [CrossRef] [PubMed]
- Podlesnykh, A.V.; Apalikova, O.V.; Brykov, V.A. Phylogenetic relationships of silver crucian carp in Carassius auratus complex based on mtDNA analysis. Russian J. Genet. 2012, 48, 1207–1217. [Google Scholar] [CrossRef]
- Wali, A.; Ahmad, S.M.; Balkhi, M.H.; Bhat, F.A.; Bhat, B.A.; Darzi, M.M. Genetic diversity of Cyprinus carpio var. communis, Cyprinus carpio var. specularis and Carassius carassius by DNA based markers. Int. J. Aquacul. 2013, 24, 138–146. [Google Scholar]
- Shen, J.B.; Fan, Z.T.; Wang, G.R. Karyotype studies of maletriploid Crucian carp (Fangzheng Crucian Carp) in Heilongjiang. J. Genet. Genom. 1983, 10, 133–136. [Google Scholar]
- Arai, R. Fish Karyotypes: A Check List; Springer: Tokyo, Japan, 2011; p. 340. [Google Scholar]
- Golubtsov, A.S.; Krysanov, E.Y. Karyological study of some cyprinid species from Ethiopia. The ploidy differences between large and small Barbus of Africa. J. Fish Biol. 1993, 42, 445–455. [Google Scholar] [CrossRef]
- Krysanov, E.Y.; Golubtsov, A.S. Karyotypes of some Ethiopian Barbus and Varicorhinus from the Nile basin including Lake Tana morphotypes. Folia Zool. 1996, 45, 67–75. [Google Scholar]
- Krysanov, E.Y. Karyotypes of Varicorhinus capoeta and Barbus goktschaicus (Cypriniformes) from lake Sevan, Armenia. J. Ichthy. 1999, 39, 262–264. [Google Scholar]
- Oellermann, L.K.; Skelton, P.H. Hexaploidy in yellowfish species (Barbus, Pisces, Cyprinidae) from southern Africa. J. Fish Biol. 1990, 37, 105–115. [Google Scholar] [CrossRef]
- Martins, C.; Galetti, P.M. Chromosomal localization of 5S rDNA gene in Leporinus fish (Anostomidae, Characiformes). Chrom. Res. 1999, 7, 363–367. [Google Scholar] [CrossRef] [PubMed]
- Inafuku, J.; Nabeyama, M.; Kikuma, Y.; Saitoh, J.; Kubota, S.; Kohno, S. Chromosomal location and nucleotide sequences of 5S ribosomal DNA of two cyprinid species (Osteichthyes, Pisces). Chrom. Res. 2000, 8, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Martins, C.; Wasko, A.P. Organization and evolution of 5S ribosomal DNA in the fish genome. In Focus on Genome Research; Williams, C.R., Ed.; Nova Science Publishers: Hauppauge, NY, USA, 2004; pp. 289–318. [Google Scholar]
- Committee for the Update of the Guide for the Care and Use of Laboratory Animals. Guide Laboratory Animals for the Care and Use of Laboratory Animals, 8th ed.; The National Academies Press: Washington, DC, USA, 2011; p. 248. [Google Scholar]
- Levan, A.; Fredgra, K.; Sandberg, A.A. Nomenclature for centromeric position on chromosomes. Hereditas 1964, 52, 201–220. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000. [Google Scholar]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef] [PubMed]
- Sevilla, R.G.; Diez, A.; Noren, M.; Mouchel, O.; Jerome, M.; Verrez-Bagnis, V.; van Pelt, H.; Favre-Krey, L.; Krey, G.; Bautista, J.M. Primers and polymerase chain reaction conditions for DNA barcoding teleost fish based on the mitochondrial cytochrome b and nuclear rhodopsin genes. Mol. Ecol. Notes 2007, 7, 730–734. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. ClustalW: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, C.-C.; Yen, T.-B.; Chen, N.-C.; Tseng, M.-C. Comparative Studies of 5S rDNA Profiles and Cyt b Sequences in two Onychostoma Species (Cyprinidae). Int. J. Mol. Sci. 2015, 16, 29663-29672. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms161226193
Han C-C, Yen T-B, Chen N-C, Tseng M-C. Comparative Studies of 5S rDNA Profiles and Cyt b Sequences in two Onychostoma Species (Cyprinidae). International Journal of Molecular Sciences. 2015; 16(12):29663-29672. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms161226193
Chicago/Turabian StyleHan, Chiao-Chuan, Tsair-Bor Yen, Nian-Cih Chen, and Mei-Chen Tseng. 2015. "Comparative Studies of 5S rDNA Profiles and Cyt b Sequences in two Onychostoma Species (Cyprinidae)" International Journal of Molecular Sciences 16, no. 12: 29663-29672. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms161226193





