Post-Translational Modification and Secretion of Azelaic Acid Induced 1 (AZI1), a Hybrid Proline-Rich Protein from Arabidopsis
Abstract
:1. Introduction
2. Results
2.1. AZI1 Retarded Mobility in Protein Gels
2.1.1. AZI1 Gel Migration Is Unrelated to MAPK Phosphorylation
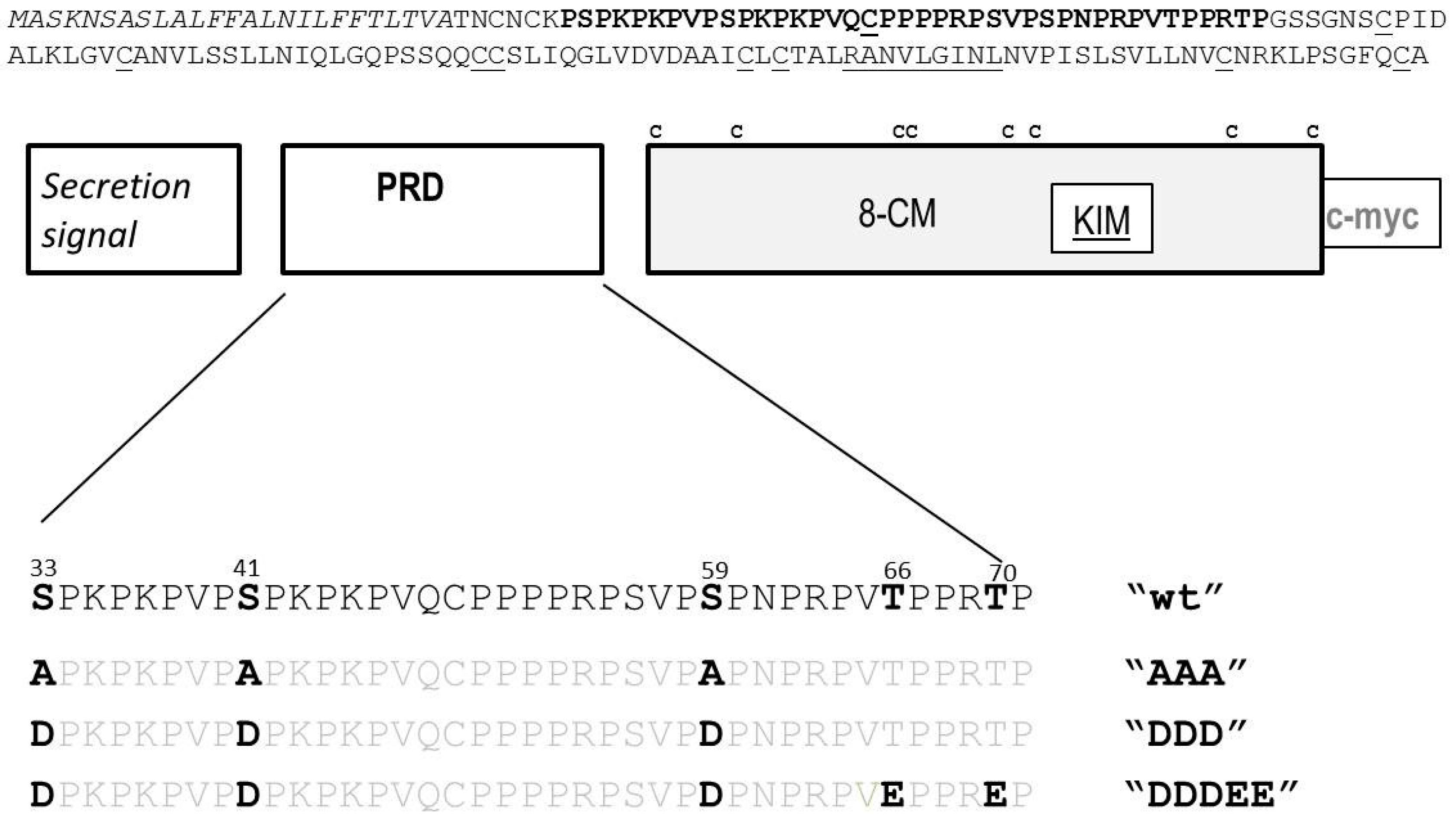
2.1.2. Substitutions at MAPK Target Sites Modify AZI1 Function
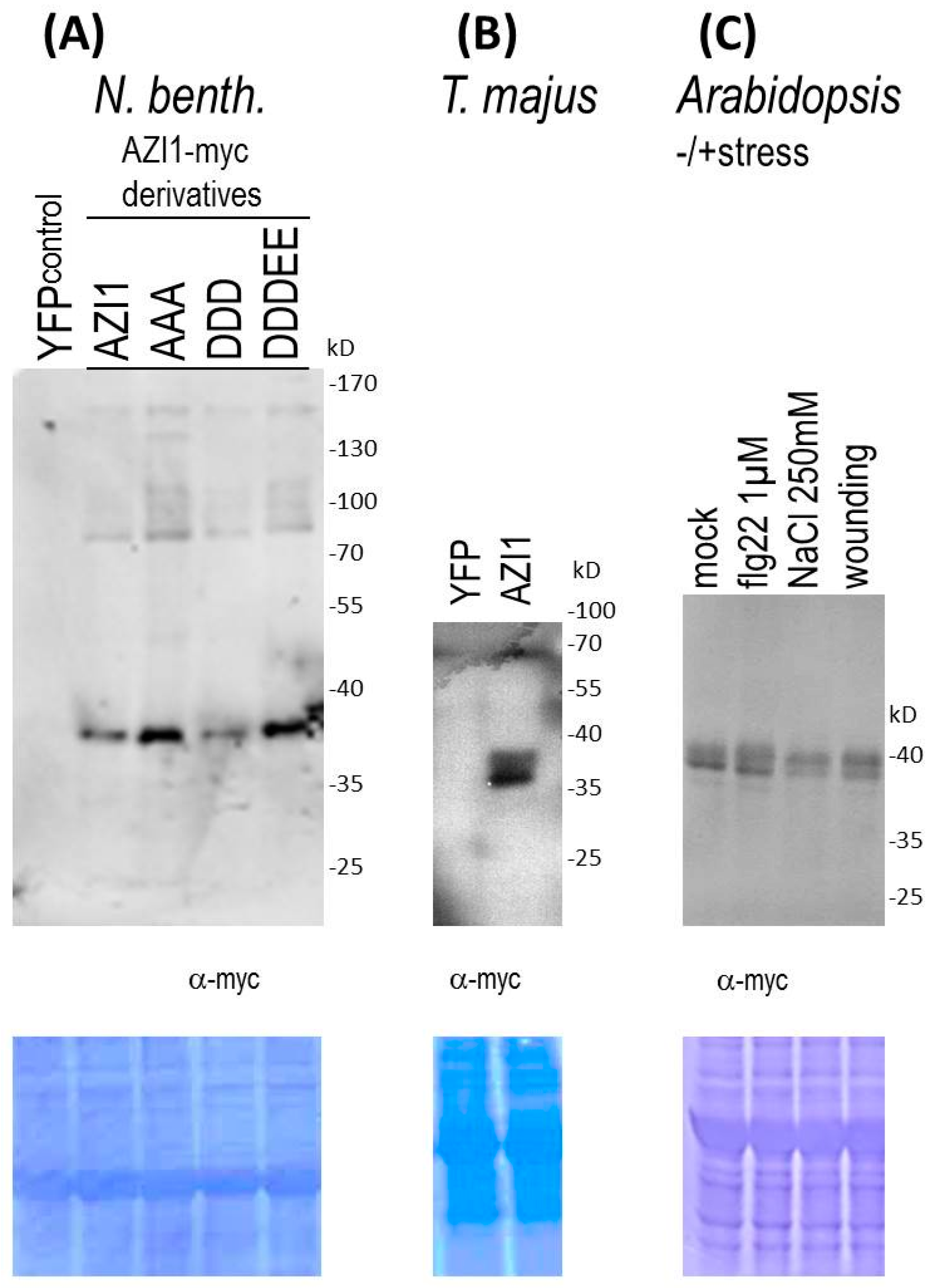
2.1.3. Indications of AZI1-Modifying Machineries in Heterologous Plant Species
2.1.4. AZI1 Gel Retardation Is Stress-Independent
2.2. O-Glycosylation Studies
2.2.1. Glycosylation—Theoretical Considerations
2.2.2. Is AZI1 an AGP?
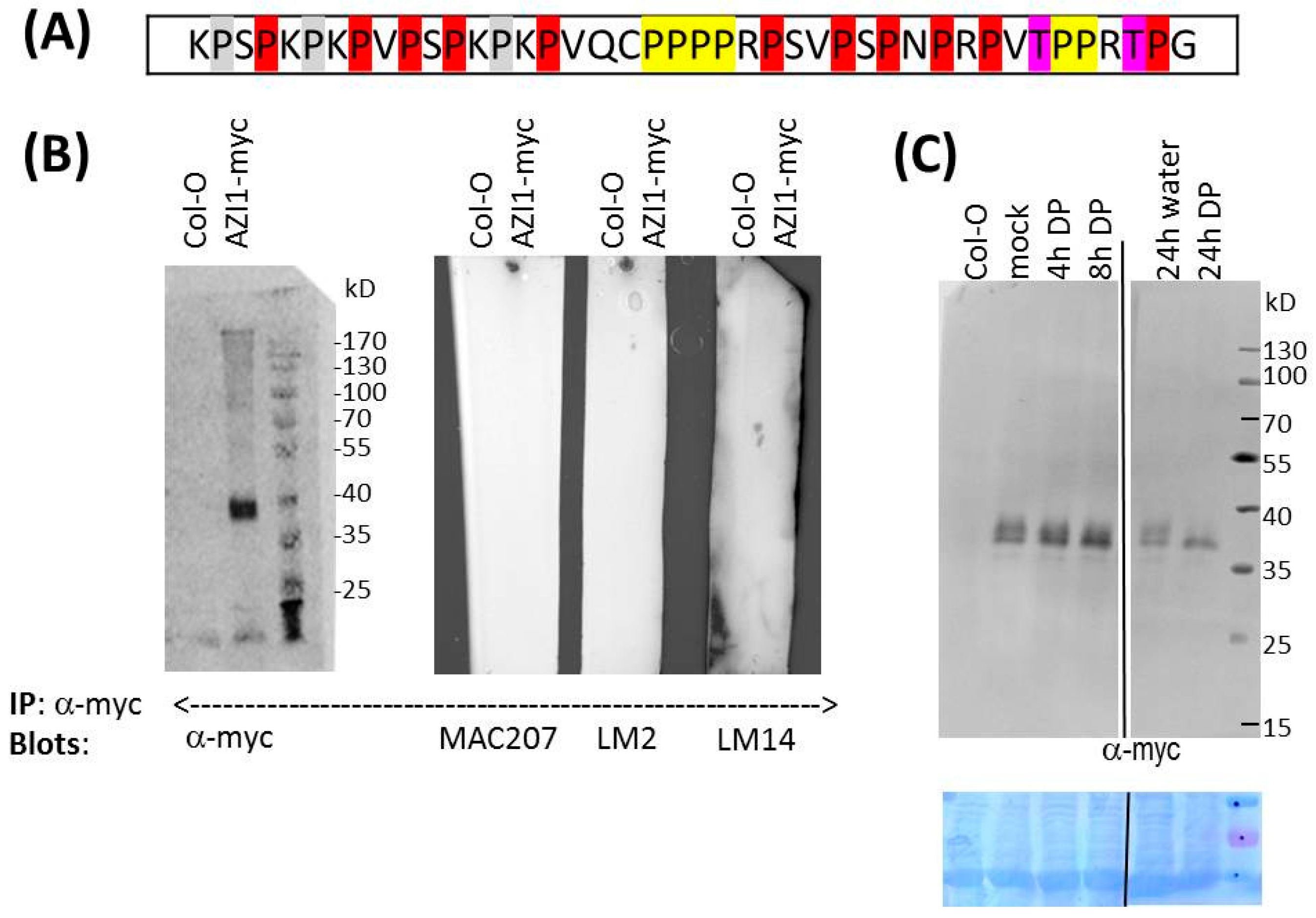
2.2.3. Pharmaceutical Approach: Inhibition of Proline Hydroxylation
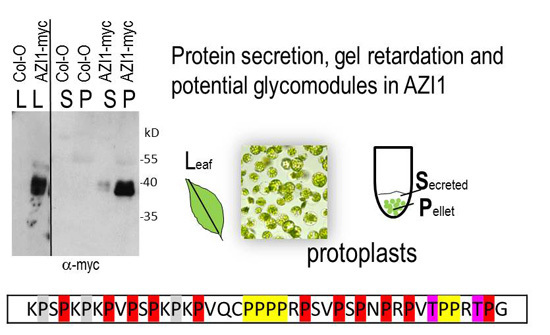
2.3. Studies on AZI1 Secretion in Vivo
2.3.1. Protoplast Secretion Studies

2.3.2. AZI1 Detection in Seedling Exudates
2.3.3. AZI1 Long-Distance Mobility in Vivo—Experimental Approaches and Limitations
3. Material and Methods
3.1. DNA Constructs and (de)-Phosphomimetic Arabidopsis Transgenic Lines
3.2. Plant Growth and Treatment
3.3. Isolation of Protoplasts and Secreted Fluid
3.4. Isolation of Apoplastic Fluid from Arabidopsis Thaliana Seedlings
3.5. Protein Extraction and Immunoblot Analysis
3.6. Immunoprecipitation
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Dvorakova, L.; Cvrckova, F.; Fischer, L. Analysis of the hybrid proline-rich protein families from seven plant species suggests rapid diversification of their sequences and expression patterns. BMC Genom. 2007, 8, 412. [Google Scholar] [CrossRef] [PubMed]
- Jose-Estanyol, M.; Gomis-Ruth, F.X.; Puigdomenech, P. The eight-cysteine motif, a versatile structure in plant proteins. Plant Physiol. Biochem. 2004, 42, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Silverstein, K.A.; Moskal, W.A., Jr.; Wu, H.C.; Underwood, B.A.; Graham, M.A.; Town, C.D.; VandenBosch, K.A. Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J. 2007, 51, 262–280. [Google Scholar] [CrossRef] [PubMed]
- Richards, K.D.; Schott, E.J.; Sharma, Y.K.; Davis, K.R.; Gardner, R.C. Aluminum induces oxidative stress genes in arabidopsis thaliana. Plant Physiol. 1998, 116, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Schlappi, M. Cold responsive earli1 type hyprps improve freezing survival of yeast cells and form higher order complexes in plants. Planta 2007, 227, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Pitzschke, A.; Datta, S.; Persak, H. Salt stress in arabidopsis: Lipid transfer protein azi1 and its control by mitogen-activated protein kinase mpk3. Mol. Plant 2014, 7, 722–738. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Zhang, X.; Xu, Z.Y.; Li, L.; Zhang, C.; Schlappi, M.; Xu, Z.Q. Influence of earli1-like genes on flowering time and lignin synthesis of arabidopsis thaliana. Plant Biol. 2011, 13, 731–739. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Huang, X.; Xu, Z.Q.; Schlappi, M. The hyprp gene earli1 has an auxiliary role for germinability and early seedling development under low temperature and salt stress conditions in arabidopsis thaliana. Planta 2011, 234, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.W.; Tschaplinski, T.J.; Wang, L.; Glazebrook, J.; Greenberg, J.T. Priming in systemic plant immunity. Science 2009, 324, 89–91. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, N.J.; Lilley, C.J.; Urwin, P.E. Identification of genes involved in the response of arabidopsis to simultaneous biotic and abiotic stresses. Plant Physiol. 2013, 162, 2028–2041. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Soares, J.M.; Mandal, M.K.; Wang, C.; Chanda, B.; Gifford, A.N.; Fowler, J.S.; Navarre, D.; Kachroo, A.; Kachroo, P. A feedback regulatory loop between g3p and lipid transfer proteins dir1 and azi1 mediates azelaic-acid-induced systemic immunity. Cell Rep. 2013, 3, 1266–1278. [Google Scholar] [CrossRef]
- Andreasson, E.; Ellis, B. Convergence and specificity in the arabidopsis mapk nexus. Trends Plant Sci. 2010, 15, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Colcombet, J.; Hirt, H. Arabidopsis mapks: A complex signalling network involved in multiple biological processes. Biochem. J. 2008, 413, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Pitzschke, A.; Schikora, A.; Hirt, H. Mapk cascade signalling networks in plant defence. Curr. Opin. Plant Biol. 2009, 12, 421–426. [Google Scholar] [CrossRef] [PubMed]
- Popescu, S.C.; Popescu, G.V.; Bachan, S.; Zhang, Z.; Gerstein, M.; Snyder, M.; Dinesh-Kumar, S.P. Mapk target networks in arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 2009, 23, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Pitzschke, A. Modes of mapk substrate recognition and control. Trends Plant Sci. 2015, 20, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Pitzschke, A.; Datta, S.; Persak, H. Mitogen-activated protein kinase-regulated azi1—An attractive candidate for genetic engineering. Plant Signal. Behav. 2014, 9, e27764. [Google Scholar] [CrossRef] [PubMed]
- Beckers, G.J.; Jaskiewicz, M.; Liu, Y.; Underwood, W.R.; He, S.Y.; Zhang, S.; Conrath, U. Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in arabidopsis thaliana. Plant Cell 2009, 21, 944–953. [Google Scholar] [CrossRef] [PubMed]
- Showalter, A.M.; Keppler, B.; Lichtenberg, J.; Gu, D.; Welch, L.R. A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol. 2010, 153, 485–513. [Google Scholar] [CrossRef] [PubMed]
- Showalter, A.M. Structure and function of plant-cell wall proteins. Plant Cell 1993, 5, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Schultz, C.J.; Johnson, K.L.; Currie, G.; Bacic, A. The classical arabinogalactan protein gene family of arabidopsis. Plant Cell 2000, 12, 1751–1768. [Google Scholar] [CrossRef] [PubMed]
- Ellis, M.; Egelund, J.; Schultz, C.J.; Bacic, A. Arabinogalactan-proteins: Key regulators at the cell surface? Plant Physiol. 2010, 153, 403–419. [Google Scholar] [CrossRef] [PubMed]
- Showalter, A.M. Arabinogalactan-proteins: Structure, expression and function. Cell. Mol. Life Sci. 2001, 58, 1399–1417. [Google Scholar] [CrossRef] [PubMed]
- Kieliszewski, M.J.; Lamport, D.T. Extensin: Repetitive motifs, functional sites, post-translational codes, and phylogeny. Plant J. 1994, 5, 157–172. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Qiu, F.; Lamport, D.T.; Kieliszewski, M.J. Structure of a hydroxyproline (hyp)-arabinogalactan polysaccharide from repetitive ala-hyp expressed in transgenic nicotiana tabacum. J. Biol. Chem. 2004, 279, 13156–13165. [Google Scholar] [CrossRef] [PubMed]
- Tryfona, T.; Liang, H.C.; Kotake, T.; Tsumuraya, Y.; Stephens, E.; Dupree, P. Structural characterization of arabidopsis leaf arabinogalactan polysaccharides. Plant Physiol. 2012, 160, 653–666. [Google Scholar] [CrossRef] [PubMed]
- Hijazi, M.; Durand, J.; Pichereaux, C.; Pont, F.; Jamet, E.; Albenne, C. Characterization of the arabinogalactan protein 31 (AGP31) of arabidopsis thaliana: New advances on the hyp-o-glycosylation of the pro-rich domain. J. Biol. Chem. 2012, 287, 9623–9632. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ngwenyama, N.; Liu, Y.; Walker, J.C.; Zhang, S. Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in arabidopsis. Plant Cell 2007, 19, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Persak, H.; Pitzschke, A. Tight interconnection and multi-level control of arabidopsis myb44 in mapk cascade signalling. PLoS ONE 2013, 8, e57547. [Google Scholar] [CrossRef] [PubMed]
- Pitzschke, A. Tropaeolum tops tobacco - simple and efficient transgene expression in the order brassicales. PLoS ONE 2013, 8, e73355. [Google Scholar] [CrossRef] [PubMed]
- Wilson, I.B. Glycosylation of proteins in plants and invertebrates. Curr. Opin. Struct. Biol. 2002, 12, 569–577. [Google Scholar] [CrossRef]
- Kieliszewski, M.J. The latest hype on hyp-o-glycosylation codes. Phytochemistry 2001, 57, 319–323. [Google Scholar] [CrossRef]
- Bardor, M.; Faye, L.; Lerouge, P. Analysis of the N-glycosylation of recombinant glycoproteins produced in transgenic plants. Trends Plant Sci. 1999, 4, 376–380. [Google Scholar] [CrossRef]
- Tryfona, T.; Liang, H.C.; Kotake, T.; Kaneko, S.; Marsh, J.; Ichinose, H.; Lovegrove, A.; Tsumuraya, Y.; Shewry, P.R.; Stephens, E.; et al. Carbohydrate structural analysis of wheat flour arabinogalactan protein. Carbohyd. Res. 2010, 345, 2648–2656. [Google Scholar] [CrossRef] [PubMed]
- Pattathil, S.; Avci, U.; Baldwin, D.; Swennes, A.G.; McGill, J.A.; Popper, Z.; Bootten, T.; Albert, A.; Davis, R.H.; Chennareddy, C.; et al. A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiol. 2010, 153, 514–525. [Google Scholar] [CrossRef] [PubMed]
- Knox, J.P. The use of antibodies to study the architecture and developmental regulation of plant cell walls. Int. Rev. Cytol. 1997, 171, 79–120. [Google Scholar] [PubMed]
- Nguema-Ona, E.; Coimbra, S.; Vicre-Gibouin, M.; Mollet, J.C.; Driouich, A. Arabinogalactan proteins in root and pollen-tube cells: Distribution and functional aspects. Ann. Bot. 2012, 110, 383–404. [Google Scholar] [CrossRef] [PubMed]
- Knox, J.P.; Linstead, P.J.; Peart, J.; Cooper, C.; Roberts, K. Developmentally regulated epitopes of cell-surface arabinogalactan proteins and their relation to root-tissue pattern-formation. Plant J. 1991, 1, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Moller, I.; Marcus, S.E.; Haeger, A.; Verhertbruggen, Y.; Verhoef, R.; Schols, H.; Ulvskov, P.; Mikkelsen, J.D.; Knox, J.P.; Willats, W. High-throughput screening of monoclonal antibodies against plant cell wall glycans by hierarchical clustering of their carbohydrate microarray binding profiles. Glycoconj. J. 2008, 25, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Speake, B.K.; Hemming, F.W.; White, D.A. The effects of tunicamycin on protein glycosylation in mammalian and fungal systems. Biochem. Soc. Trans. 1980, 8, 166–168. [Google Scholar] [CrossRef] [PubMed]
- Barnett, N.M. Dipyridyl-induced cell elongation and inhibition of cell wall hydroxyproline biosynthesis. Plant Physiol. 1970, 45, 188–191. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Calvino, L.; Faulkner, C.; Walshaw, J.; Saalbach, G.; Bayer, E.; Benitez-Alfonso, Y.; Maule, A. Arabidopsis plasmodesmal proteome. PLoS ONE 2011, 6, e18880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitra, S.K.; Walters, B.T.; Clouse, S.D.; Goshe, M.B. An efficient organic solvent based extraction method for the proteomic analysis of arabidopsis plasma membranes. J. Proteom. Res. 2009, 8, 2752–2767. [Google Scholar] [CrossRef] [PubMed]
- Young, B.; Wightman, R.; Blanvillain, R.; Purcel, S.B.; Gallois, P. pH-sensitivity of YFP provides an intracellular indicator of programmed cell death. Plant Method 2010, 6, 27. [Google Scholar] [CrossRef] [PubMed]
- Griesbeck, O.; Baird, G.S.; Campbell, R.E.; Zacharias, D.A.; Tsien, R.Y. Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J. Biol. Chem. 2001, 276, 29188–29194. [Google Scholar] [CrossRef] [PubMed]
- Hellens, R.P.; Edwards, E.A.; Leyland, N.R.; Bean, S.; Mullineaux, P.M. Pgreen: A versatile and flexible binary ti vector for agrobacterium-mediated plant transformation. Plant Mol. Biol. 2000, 42, 819–832. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for agrobacterium-mediated transformation of arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Pitzschke, A.; Persak, H. Poinsettia protoplasts—A simple, robust and efficient system for transient gene expression studies. Plant Methods 2012, 8, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, F.H.; Shen, S.C.; Lee, L.Y.; Lee, S.H.; Chan, M.T.; Lin, C.S. Tape-arabidopsis sandwich—A simpler arabidopsis protoplast isolation method. Plant Methods 2009, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Shpak, E.; Leykam, J.F.; Kieliszewski, M.J. Synthetic genes for glycoprotein design and the elucidation of hydroxyproline-o-glycosylation codes. Proc. Natl. Acad. Sci. USA 1999, 96, 14736–14741. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Zhao, J. Genome-wide identification, classification, and expression analysis of the arabinogalactan protein gene family in rice (Oryza sativa L.). J. Exp. Bot. 2010, 61, 2647–2668. [Google Scholar] [CrossRef] [PubMed]
- Motose, H.; Sugiyama, M.; Fukuda, H. A proteoglycan mediates inductive interaction during plant vascular development. Nature 2004, 429, 873–878. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, T.C.; Domingo, C.; Schindler, T.; Seetharaman, G.; Stacey, N.; Roberts, K. Dcagp1, a secreted arabinogalactan protein, is related to a family of basic proline-rich proteins. Plant Mol. Biol. 2001, 45, 421–435. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.L.; Jones, B.J.; Bacic, A.; Schultz, C.J. The fasciclin-like arabinogalactan proteins of arabidopsis. A multigene family of putative cell adhesion molecules. Plant Physiol. 2003, 133, 1911–1925. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pitzschke, A.; Xue, H.; Persak, H.; Datta, S.; Seifert, G.J. Post-Translational Modification and Secretion of Azelaic Acid Induced 1 (AZI1), a Hybrid Proline-Rich Protein from Arabidopsis. Int. J. Mol. Sci. 2016, 17, 85. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17010085
Pitzschke A, Xue H, Persak H, Datta S, Seifert GJ. Post-Translational Modification and Secretion of Azelaic Acid Induced 1 (AZI1), a Hybrid Proline-Rich Protein from Arabidopsis. International Journal of Molecular Sciences. 2016; 17(1):85. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17010085
Chicago/Turabian StylePitzschke, Andrea, Hui Xue, Helene Persak, Sneha Datta, and Georg J. Seifert. 2016. "Post-Translational Modification and Secretion of Azelaic Acid Induced 1 (AZI1), a Hybrid Proline-Rich Protein from Arabidopsis" International Journal of Molecular Sciences 17, no. 1: 85. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17010085