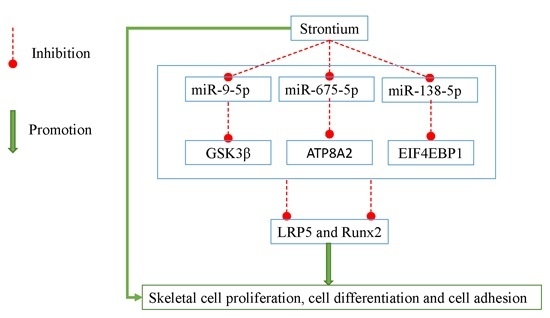
miR-9-5p, miR-675-5p and miR-138-5p Damages the Strontium and LRP5-Mediated Skeletal Cell Proliferation, Differentiation, and Adhesion
Abstract
:1. Introduction
2. Results
2.1. Strontium Altered the Expression Levels of miRNAs in Vitro and in Vivo

2.2. miR-9-5p, miR-675-5p, and miR-138-5p Reduced Skeletal Cell Count and Cell Proliferation

2.3. miR-9-5p, miR-675-5p, and miR-138-5p Impaired Skeletal Cell Differentiation and Cell Adhesion

2.4. miR-9-5p and miR-675-5p Induced MC3T3-E1 Cell Apoptosis More Specifically than miR-138-5p

2.5. miR-9-5p, miR-675-5p, and miR-138-5p Targeted GSK3β, ATP8A2, and EIF4EBP1

2.6. LRP5 Played Positive Roles in Skeletal Development

3. Discussion

4. Experimental Section
4.1. Cell Cultures and Reagents
4.2. Plasmid Construction
4.3. Transfection
4.4. Clonogenic Assay (Colony Formation Assay)
4.5. Cell Viability Assay
4.6. Cell Cytotoxicity Assay
4.7. TUNEL Cell Apoptosis Assay
4.8. Animal
4.9. RNA Extraction
4.10. Poly (A) Polymerase Tailing
4.11. Reverse Transcription
4.12. Quantitative Real-Time PCR (qPCR)
| Primer Name | Sequences (5′–3′) |
|---|---|
| RACE Reverse | GCGAGCACAGAATTAATACGAC |
| miR-9-5p Forward | GCGTCTTTGGTTATCTAGCTGTA |
| miR-675-5p Forward | TGGTGCGGAAAGGGCCCACAGT |
| miR-138-5p Forward | AGCTGGTGTTGTGAATCAGGCCG |
| miR-370 Forward | GCCTGCTGGGGTGGAACCTGGT |
| miR-23a Forward | ATCACATTGCCAGGGATTTCC |
| miR-103 Forward | ACACTCCAGCTGGGAGCAGCATTGTAC |
| GAPDH Forward | ATTGTCAGCAATGCATCCTG |
| GAPDH Reverse | ATGGACTGTGGTCATGAGCC |
| ALP Forward | AACCCAGACACAAGCATTCC |
| ALP Reverse | GCCTTTGAGGTTTTTGGTCA |
| COL1A1 Forward | GCCAAGAAGACATCCCTGAA |
| COL1A1 Reverse | GCCATTGTGGCAGATACAGA |
| RUNX2 Forward | AAGTGCGGTGCAAACTTTCT |
| RUNX2 Reverse | ACGCCATAGTCCCTCCTTTT |
| GSK3β Forward | TTGGACAAAGGTCTTCCGGC |
| GSK3β Reverse | AAGAGTGCAGGTGTGTCTCG |
| ATP8A2 Forward | ACGAGGGACGTGCTCATGAAGC |
| ATP8A2 Reverse | CCTCAAGTGTCACCAGCAGGCT |
| EIF4EBP1 Forward | CTAGCCCTACCAGCGATGAG |
| EIF4EBP1 Reverse | CCTGGTATGAGGCCTGAATG |
| LRP5 Forward | TGCCACTGGTGAGATTGAC |
| LRP5 Reverse | ACTGCTGCTTGATGAGGAC |
| Cyclin-D1 Forward | GCGTACCCTGACACCAATCT |
| Cyclin-D1 Reverse | CTCCTCTTCGCACTTCTGCT |
| CyclinA Forward | CAGAGGCCGAAGACGAGAC |
| CyclinA Reverse | TCAGCTGGCTTCTTCTGAGC |
| CyclinE Forward | GTTATAAGGGAGACGGGGAG |
| CyclinE Reverse | TGCTCTGCTTCTTACCGCTC |
| PCNA Forward | TTTGAGGCACGCCTGATCC |
| PCNA Reverse | GGAGACGTGAGACGAGTCCAT |
| Integrin α2 Forward | AAGTGCCCTGTGGACCTACCCA |
| Integrin α2 Reverse | TGGTGAGGGTCAATCCCAGGCT |
| Integrin α5 Forward | ACCACCTGCAGAAACGAGAGGC |
| Integrin α5 Reverse | TGGCCCAAACTCACAGCGCA |
| Integrin Αv Forward | TCCCACCGCAGGCTGACTTCAT |
| Integrin Αv Reverse | TCGGGTTTCCAAGGTCGCACAC |
| Integrin β1 Forward | TTCAGACTTCCGCATTGGCT |
| Integrin β1 Reverse | AATGGGCTGGTGCAGTTTTG |
| Fibronectin Forward | TGCAGTGGCTGAAGTCGCAAGG |
| Fibronectin Reverse | GGGCTCCCCGTTTGAATTGCCA |
| FAK Forward | AGCACCTGGCCACCTAAGCAAC |
| FAK Reverse | CATTGGACCGGTCAAGGTTGGCA |
| Vinculin Forward | TCAAGCTGTTGGCAGTAGCCGC |
| Vinculin Reverse | TCTCTGCTGTGGCTCCAAGCCT |
| Osteocalcin Forward | AGCAGGAGGGCAATAAGGTAGT |
| Osteocalcin Reverse | TCGTCACAAGCAGGGTTAAGC |
| Osteonectin Forward | ATGTCCTGGTCACCTTGTACGA |
| Osteonectin Reverse | TCCAGGCGCTTCTCATTCTCAT |
| Osteopontin Forward | TGATTCTGGCAGCTCAGAGGA |
| Osteopontin Reverse | CATTCTGTGGCGCAAGGAGATT |
| Type I collagen Forward | CTCCTGACGCATGGCCAAGAA |
| Type I collagen Reverse | TCAAGCATACCTCGGGTTTCCA |
| Type III collagen Forward | CCCTGGCTCAAATGGCTCACCA |
| Type III collagen Reverse | CCTTTCCACCAGGACTGCCGTT |
| Cadherin 11 Forward | GGCCCAAACAGGTATCATCA |
| Cadherin 11 Reverse | TTGGTTGTCCCTGAGAGTCC |
| MAPK1 Forward | ATCCGGGCACCAACCATTGAGC |
| MAPK1 Reverse | GTGGTCATTGCTGAGGTGCTGTGT |
| NFkB Forward | AACCCATCGCCTTGGCATCCAC |
| NFkB Reverse | AGTCGAAAAGGGCGTTGGCGT |
| FasL Forward | GAGAATTGCTGAAGACATGACAATCC |
| FasL Reverse | ATGGCTGGAACTGAGGTAGTTTTCAC |
| p53 Forward | TAACAGTTCCTGCATGGGCGGC |
| p53 Reverse | CGGAGGCCCATCCTCACCATCATCA |
| HEY1 Forward | GCATACGGCAGGAGGGAAA |
| HEY1 Reverse | CTGGGAAGCGTAGTTGTTGAGAT |
| Sost Forward | GGAATGATGCCACAGAGGTCAT |
| Sost Reverse | CCCGGTTCATGGTCTGGTT |
| Smad7 Forward | GGACGCTGTTGGTACACAAG |
| Smad7 Reverse | GCTGCATAAACTCGTGGTCATTG |
| SMAD6 Forward | GCTCTAGGAATGCAGACGCTG |
| SMAD6 Reverse | CAACAGGCAGTCAGCACAGTC |
4.13. Luciferase Reporter Assay
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Saidak, Z.; Marie, P.J. Strontium signaling: Molecular mechanisms and therapeutic implications in osteoporosis. Pharmacol. Ther. 2012, 136, 216–226. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Zhou, G.; Luk, K.D.; Cheung, K.M.; Li, Z.; Lam, W.M.; Zhou, Z.; Lu, W.W. Strontium promotes osteogenic differentiation of mesenchymal stem cells through the Ras/MAPK signaling pathway. Cell. Physiol. Biochem. 2009, 23, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Qu, B.; Xia, X.; Wu, H.H.; Tu, C.Q.; Pan, X.M. PDGF-regulated miRNA-138 inhibits the osteogenic differentiation of mesenchymal stem cells. Biochem. Biophys. Res. Commun. 2014, 448, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Itoh, T.; Ando, M.; Tsukamasa, Y.; Akao, Y. Expression of BMP-2 and Ets1 in BMP-2-stimulated mouse pre-osteoblast differentiation is regulated by microRNA-370. FEBS Lett. 2012, 586, 1693–1701. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.Y.; Lee, H.F.V.; Wong, A.M.; Kwong, D.L.; Zhu, Y.H.; Dong, S.S.; Kong, K.L.; Chen, J.; Tsao, S.W.; Guan, X.Y.; et al. MicroRNA-144 promotes cell proliferation, migration and invasion in nasopharyngeal carcinoma through repression of PTEN. Carcinogenesis 2013, 34, 454–463. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Liu, X.S.; Zhou, G.; Li, Z.; Luk, K.D.; Guo, X.E.; Lu, W.W. Osteoprotegerin deficiency attenuates strontium-mediated inhibition of osteoclastogenesis and bone resorption. J. Bone Miner. Res. 2011, 26, 1272–1282. [Google Scholar] [CrossRef] [PubMed]
- Thouverey, C.; Caverzasio, J. Sclerostin inhibits osteoblast differentiation without affecting BMP2/SMAD1/5 or Wnt3a/β-catenin signaling but through activation of platelet-derived growth factor receptor signaling in vitro. BoneKEY Rep. 2015, 4, 757. [Google Scholar] [CrossRef] [PubMed]
- Ellies, D.L.; Viviano, B.; McCarthy, J.; Rey, J.P.; Itasaki, N.; Saunders, S.; Krumlauf, R. Bone density ligand, Sclerostin, directly interacts with LRP5 but not LRP5G171V to modulate Wnt activity. J. Bone Miner. Res. 2006, 21, 1738–1749. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, Y.; Kang, H.; Liu, W.; Liu, P.; Zhang, J.; Harris, S.E.; Wu, D. Sclerostin binds to LRP5/6 and antagonizes canonical Wnt signaling. J. Biol. Chem. 2005, 280, 19883–19887. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Yin, X.; Zhang, X.; Jing, D.; Feng, X. Cyclic stretch enhances bone morphogenetic protein-2-induced osteoblastic differentiation through the inhibition of Hey1. Int. J. Mol. Med. 2015, 36, 1273–1281. [Google Scholar] [CrossRef] [PubMed]
- Yano, M.; Inoue, Y.; Tobimatsu, T.; Hendy, G.; Canaff, L.; Sugimoto, T.; Seino, S.; Kaji, H. Smad7 inhibits differentiation and mineralization of mouse osteoblastic cells. Endocr. J. 2012, 59, 653–662. [Google Scholar] [CrossRef] [PubMed]
- Min, Z.; Zhang, R.; Yao, J.; Jiang, C.; Guo, Y.; Cong, F.; Wang, W.; Tian, J.; Zhong, N.; Sun, J.; Ma, J.; Lu, S. MicroRNAs associated with osteoarthritis differently expressed in bone matrix gelatin (BMG) rat model. Int. J. Clin. Exp. Med. 2015, 8, 1009–1017. [Google Scholar] [PubMed]
- Wang, S.; Tang, C.; Zhang, Q.; Chen, W. Reduced miR-9 and miR-181a expression down-regulates Bim concentration and promote osteoclasts survival. Int. J. Clin. Exp. Pathol. 2014, 7, 2209–2218. [Google Scholar] [PubMed]
- Huang, Y.; Zheng, Y.; Jia, L.; Li, W. Long noncoding RNA H19 promotes osteoblast differentiation via TGF-β1/Smad3/HDAC signaling pathway by deriving miR-675. Stem Cells 2015, 33, 3481–3492. [Google Scholar] [CrossRef] [PubMed]
- Martin, P.J.; Haren, N.; Ghali, O.; Clabaut, A.; Chauveau, C.; Hardouin, P.; Broux, O. Adipogenic RNAs are transferred in osteoblasts via bone marrow adipocytes-derived extracellular vesicles (EVs). BMC Cell Biol. 2015, 16, 10. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.L.; Liu, H.Z.; Zhang, Z.C.; Zhao, H.L.; Zhao, H.; Huang, Y.; Yao, J.H.; Sun, T.S. The influence of MicroRNA-150 in Osteoblast Matrix Mineralization. J. Cell. Biochem. 2015, 116, 2970–2979. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Cui, X.; Jiang, Z.; Sun, J. MicroRNA-23a modulates tumor necrosis factor-α-induced osteoblasts apoptosis by directly targeting Fas. J. Cell. Biochem. 2013, 114, 2738–2745. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Cao, X.; Hu, Z.; Zhang, L.; Wang, H.; Zhou, H.; Li, D.; Zhang, S.; Xie, M. MiR-103 inhibits osteoblast proliferation mainly through suppressing Cav1.2 expression in simulated microgravity. Bone 2015, 76, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Cheng, J.; Zeng, Z.; Wang, Y.; Li, X.; Xie, Q.; Jia, J.; Yan, Y.; Guo, Z.; Gao, J.; et al. Comprehensive profiling of novel microRNA-9 targets and a tumor suppressor role of microRNA-9 via targeting IGF2BP1 in hepatocellular carcinoma. Oncotarget 2015, 6, 42040–42052. [Google Scholar] [PubMed]
- Sun, Z.; Han, Q.; Zhou, N.; Wang, S.; Lu, S.; Bai, C.; Zhao, R.C. MicroRNA-9 enhances migration and invasion through KLF17 in hepatocellular carcinoma. Mol. Oncol. 2013, 7, 884–894. [Google Scholar] [CrossRef] [PubMed]
- Sista, S.; Wen, C.; Hodgson, P.D.; Pande, G. Expression of cell adhesion and differentiation related genes in MC3T3 osteoblasts plated on titanium alloys: Role of surface properties. Mater. Sci. Eng. C Mater. Biol. Appl. 2013, 33, 1573–1582. [Google Scholar] [CrossRef] [PubMed]
- Kahai, S.; Lee, S.C.; Lee, D.Y.; Yang, J.; Li, M.; Wang, C.H.; Jiang, Z.; Zhang, Y.; Peng, C.; Yang, B.B. MicroRNA miR-378 regulates nephronectin expression modulating osteoblast differentiation by targeting GalNT-7. PLoS ONE 2009, 4, e7535. [Google Scholar] [CrossRef] [PubMed]
- Hamam, D.; Ali, D.; Vishnubalaji, R.; Hamam, R.; Al-Nbaheen, M.; Chen, L.; Kassem, M.; Aldahmash, A.; Alajez, N.M. microRNA-320/RUNX2 axis regulates adipocytic differentiation of human mesenchymal (skeletal) stem cells. Cell Death Dis. 2014, 5, e1499. [Google Scholar] [CrossRef] [PubMed]
- Wei, N.; Xiao, L.; Xue, R.; Zhang, D.; Zhou, J.; Ren, H.; Guo, S.; Xu, J. MicroRNA-9 mediates the cell apoptosis by targeting Bcl2l11 in ischemic stroke. Mol. Neurobiol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Mei, G.; Zou, Z.; Fu, S.; Xia, L.; Zhou, J.; Zhang, Y.; Tuo, Y.; Wang, Z.; Jin, D. Substance P activates the Wnt signal transduction pathway and enhances the differentiation of mouse preosteoblastic MC3T3-E1 cells. Int. J. Mol. Sci. 2014, 15, 6224–6240. [Google Scholar] [CrossRef] [PubMed]
- Kohno, T.; Tsuge, M.; Murakami, E.; Hiraga, N.; Abe, H.; Miki, D.; Imamura, M.; Ochi, H.; Hayes, C.N.; Chayama, K. Human microRNA hsa-miR-1231 suppresses hepatitis B virus replication by targeting core mRNA. J. Viral Hepat. 2014, 21, e89–e97. [Google Scholar] [CrossRef] [PubMed]
- Franken, N.A.; Rodermond, H.M.; Stap, J.; Haveman, J.; van Bree, C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006, 1, 2315–2319. [Google Scholar] [CrossRef] [PubMed]
- Reichenstein, I.; Aizenberg, N.; Goshen, M.; Bentwich, Z.; Avni, Y.S. A novel qPCR assay for viral encoded microRNAs. J. Virol. Methods 2010, 163, 323–328. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, T.; Leung, F.; Lu, W.W. miR-9-5p, miR-675-5p and miR-138-5p Damages the Strontium and LRP5-Mediated Skeletal Cell Proliferation, Differentiation, and Adhesion. Int. J. Mol. Sci. 2016, 17, 236. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17020236
Sun T, Leung F, Lu WW. miR-9-5p, miR-675-5p and miR-138-5p Damages the Strontium and LRP5-Mediated Skeletal Cell Proliferation, Differentiation, and Adhesion. International Journal of Molecular Sciences. 2016; 17(2):236. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17020236
Chicago/Turabian StyleSun, Tianhao, Frankie Leung, and William W. Lu. 2016. "miR-9-5p, miR-675-5p and miR-138-5p Damages the Strontium and LRP5-Mediated Skeletal Cell Proliferation, Differentiation, and Adhesion" International Journal of Molecular Sciences 17, no. 2: 236. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17020236