G-Protein Dependent Signal Transduction and Ubiquitination in Dictyostelium
Abstract
:1. Introduction
2. The Dictyostelium Ubiquitin Landscape: A Short Glance
3. G-Protein Coupled Receptors and Their Molecular Switches
4. Navigating Downstream of G-Protein: Ras, PI3K and TORC2
5. Intracellular cAMP Signalling Converges to PKA
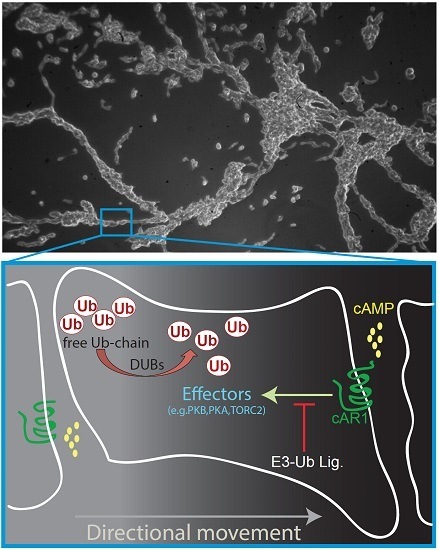
6. Is cAR1 Dependent Signalling Pathway Regulated by Ubiquitination?
7. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Yau, R.; Rape, M. The increasing complexity of the ubiquitin code. Nat. Cell Biol. 2016, 27, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Schwartz, D.; Elias, J.E.; Thoreen, C.C.; Cheng, D.; Marsischky, G.; Roelofs, J.; Finley, D.; Gygi, S.P. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 2003, 21, 921–926. [Google Scholar] [CrossRef] [PubMed]
- Artemenko, Y.; Lampert, T.J.; Devreotes, P.N. Moving towards a paradigm: Common mechanisms of chemotactic signaling in dictyostelium and mammalian leukocytes. Cell. Mol. Life Sci. 2014, 71, 3711–3747. [Google Scholar] [CrossRef] [PubMed]
- Kessin, R.H. The evolution of the cellular slime molds. In Dictyostelium—A Model System for Cell and Developmental Biology; Maeda, Y., Inouye, K., Takeuchi, I., Eds.; Universal Academy Press: Tokyo, Japan, 1997; pp. 3–13. [Google Scholar]
- De Lozanne, A. Homologous recombination in dictyostelium as a tool for the study of developmental genes. Meth. Cell Biol. 1987, 28, 489–495. [Google Scholar]
- Kuspa, A.; Loomis, W.F. Tagging developmental genes in dictyostelium by restriction enzyme-mediated integration of plasmid DNA. Proc. Natl. Acad. Sci. USA 1992, 89, 8803–8807. [Google Scholar] [CrossRef] [PubMed]
- Eichinger, L.; Pachebat, J.A.; Glockner, G.; Rajandream, M.A.; Sucgang, R.; Berriman, M.; Song, J.; Olsen, R.; Szafranski, K.; Xu, Q.; et al. The genome of the social amoeba dictyostelium discoideum. Nature 2005, 435, 43–57. [Google Scholar] [CrossRef] [PubMed]
- Bozzaro, S. The model organism dictyostelium discoideum. Methods Mol. Biol. 2013, 983, 17–37. [Google Scholar] [PubMed]
- Devreotes, P.; Horwitz, A.R. Signaling networks that regulate cell migration. Cold Spring Harb. Perspect. Biol. 2015, 7, a005959. [Google Scholar] [CrossRef] [PubMed]
- Annesley, S.J.; Chen, S.; Francione, L.M.; Sanislav, O.; Chavan, A.J.; Farah, C.; De Piazza, S.W.; Storey, C.L.; Ilievska, J.; Fernando, S.G.; et al. Dictyostelium, a microbial model for brain disease. Biochim. Biophys. Acta 2014, 1840, 1413–1432. [Google Scholar] [CrossRef] [PubMed]
- Bracco, E.; Pergolizzi, B. Ras proteins signaling in the early metazoan dictyostelium discoideum. Methods Mol. Biol. 2014, 1120, 407–420. [Google Scholar] [PubMed]
- Weissman, A.M. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2001, 2, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Gonzalo, F.R.; Rosa, J.L. The herc proteins: Functional and evolutionary insights. Cell. Mol. Life Sci. 2005, 62, 1826–1838. [Google Scholar] [CrossRef] [PubMed]
- Pickart, C.M.; Eddins, M.J. Ubiquitin: Structures, functions, mechanisms. Biochim. Biophys. Acta 2004, 29, 55–72. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Pickart, C.M. Different hect domain ubiquitin ligases employ distinct mechanisms of polyubiquitin chain synthesis. Embo J 2005 2005, 21, 4324–4333. [Google Scholar] [CrossRef] [PubMed]
- Buetow, L.; Huang, D.T. Structural insights into the catalysis and regulation of e3 ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2016, 17, 626–642. [Google Scholar] [CrossRef] [PubMed]
- Kulathu, Y.; Komander, D. Atypical ubiquitylation—The unexplored world of polyubiquitin beyond lys48 and lys63 linkages. Nat. Rev. Mol. Cell Biol. 2012, 13, 508–523. [Google Scholar] [CrossRef] [PubMed]
- Nijman, S.M.; Luna-Vargas, M.P.; Velds, A.; Brummelkamp, T.R.; Dirac, A.M.; Sixma, T.K.; Bernards, R. A genomic and functional inventory of deubiquitinating enzymes. Cell 2005, 123, 773–786. [Google Scholar] [CrossRef] [PubMed]
- Ohmachi, T.; Giorda, R.; Shaw, D.R.; Ennis, H.L. Molecular organization of developmentally regulated dictyostelium discoideum ubiquitin cdnas. Biochemistry 1989, 28, 5226–5231. [Google Scholar] [CrossRef] [PubMed]
- Muller-Taubenberger, A.; Hagmann, J.; Noegel, A.; Gerisch, G. Ubiquitin gene expression in dictyostelium is induced by heat and cold shock, cadmium, and inhibitors of protein synthesis. J. Cell Sci. 1988, 90, 51–58. [Google Scholar] [PubMed]
- Schauer, T.M.; Nesper, M.; Kehl, M.; Lottspeich, F.; Muller-Taubenberger, A.; Gerisch, G.; Baumeister, W. Proteasomes from dictyostelium discoideum: Characterization of structure and function. J. Struct. Biol. 1993, 111, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Masson, P.; Lundin, D.; Soderbom, F.; Young, P. Characterization of a reg/pa28 proteasome activator homolog in dictyostelium discoideum indicates that the ubiquitin- and atp-independent reggamma proteasome is an ancient nuclear protease. Eukaryot. Cell 2009, 8, 844–851. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, D.F.; Amerik, A.; Deery, W.J.; Bishop, J.D.; Hochstrasser, M.; Gomer, R.H. A deubiquitinating enzyme that disassembles free polyubiquitin chains is required for development but not growth in dictyostelium. J. Biol. Chem. 1998, 273, 29178–29187. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.Y.; Reddy, T.B.K.; Zhou, K.M.; Firtel, R.A. A novel, putative mek kinase controls developmental timing and spatial patterning in dictyostelium and is regulated by ubiquitin-mediated protein degradation. Genes Dev. 1998, 12, 3564–3578. [Google Scholar] [CrossRef] [PubMed]
- Clark, A.; Nomura, A.; Mohanty, S.; Firtel, R.A. A ubiquitin-conjugating enzyme is essential for developmental transitions in dictyostelium. Mol. Biol. Cell 1997, 8, 1989–2002. [Google Scholar] [CrossRef] [PubMed]
- Langenick, J.; Araki, T.; Yamada, Y.; Williams, J.G. A dictyostelium homologue of the metazoan Cbl proteins regulates stat signalling. J. Cell Sci. 2008, 121, 3524–3530. [Google Scholar] [CrossRef] [PubMed]
- Sobko, A.; Ma, H.; Firtel, R.A. Regulated SUMOylation and ubiquitination of DdMEK1 is required for proper chemotaxis. Dev. Cell 2002, 2, 745–756. [Google Scholar] [CrossRef]
- Mohanty, S.; Lee, S.; Yadava, N.; Dealy, M.J.; Johnson, R.S.; Firtel, R.A. Regulated protein degradation controls pka function and cell-type differentiation in dictyostelium. Genes Dev. 2001, 15, 1435–1448. [Google Scholar] [CrossRef] [PubMed]
- Ennis, H.L.; Dao, D.N.; Pukatzki, S.U.; Kessin, R.H. Dictyostelium amoebae lacking an f-box protein form spores rather than stalk in chimeras with wild type. Proc. Natl. Acad. Sci. USA 2000, 97, 3292–3297. [Google Scholar] [CrossRef] [PubMed]
- Whitney, N.; Pearson, L.J.; Lunsford, R.; McGill, L.; Gomer, R.H.; Lindsey, D.F. A putative ariadne-like ubiquitin ligase is required for dictyostelium discoideum development. Eukaryot. Cell 2006, 5, 1820–1825. [Google Scholar] [CrossRef] [PubMed]
- Blagg, S.L.; Battom, S.E.; Annesley, S.J.; Keller, T.; Parkinson, K.; Wu, J.M.; Fisher, P.R.; Thompson, C.R. Cell type-specific filamin complex regulation by a novel class of hect ubiquitin ligase is required for normal cell motility and patterning. Development 2011, 138, 1583–1593. [Google Scholar] [CrossRef] [PubMed]
- Pergolizzi, B.; Bracco, E.; Bozzaro, S. A new hect ubiquitin ligase regulating chemotaxis and development in dictyostelium discoideum. J. Cell Sci. 2017, 130, 551–562. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, Y.; Eichinger, L. The dictyostelium repertoire of seven transmembrane domain receptors. Eur. J. Cell Biol. 2006, 85, 937–946. [Google Scholar] [CrossRef] [PubMed]
- Pan, M.; Xu, X.; Chen, Y.; Jin, T. Identification of a chemoattractant g-protein-coupled receptor for folic acid that controls both chemotaxis and phagocytosis. Dev. Cell 2016, 36, 428–439. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, Y.; Mondal, S.; Eichinger, L.; Noegel, A.A. A gpcr involved in post aggregation events in dictyostelium discoideum. Dev. Biol. 2007, 312, 29–43. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, Y.; Muller, R.; Anjard, C.; Noegel, A.A. Grlj, a dictyostelium gabab-like receptor with roles in post-aggregation development. BMC Dev. Biol. 2007, 7, 44. [Google Scholar] [CrossRef] [PubMed]
- Robery, S.; Tyson, R.; Dinh, C.; Kuspa, A.; Noegel, A.A.; Bretschneider, T.; Andrews, P.L.; Williams, R.S. A novel human receptor involved in bitter tastant detection identified using dictyostelium discoideum. J. Cell Sci. 2013, 126, 5465–5476. [Google Scholar] [CrossRef] [PubMed]
- Hereld, D.; Devreotes, P.N. The camp receptor family of dictyostelium. Int. Rev. Cytol. 1992, 137B, 35–47. [Google Scholar] [PubMed]
- Raisley, B.; Zhang, M.H.; Hereld, D.; Hadwiger, J.A. A camp receptor-like g protein-coupled receptor with roles in growth regulation and development. Dev. Biol. 2004, 265, 433–445. [Google Scholar] [CrossRef] [PubMed]
- Hadwiger, J.A. Developmental morphology and chemotactic responses are dependent on galpha subunit specificity in dictyostelium. Dev. Biol. 2007, 312, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.J.; Valkema, R.; van Haastert, P.J.M.; Devreotes, P.N. The g protein beta subunit is essential for multiple responses to chemoattractants in dictyostelium. J. Cell Biol. 1995, 129, 1667–1675. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Long, Y.; Devreotes, P.N. Ggamma in dictyostelium: Its role in localization of gbetagamma to the membrane is required for chemotaxis in shallow gradients. Mol. Biol. Cell 2001, 12, 3204–3213. [Google Scholar] [CrossRef] [PubMed]
- Brzostowski, J.A.; Parent, C.A.; Kimmel, A.R. A galpha-dependent pathway that antagonizes multiple chemoattractant responses that regulate directional cell movement. Genes Dev. 2004, 18, 805–815. [Google Scholar] [CrossRef] [PubMed]
- Hereld, D.; Vaughan, R.; Kim, J.Y.; Borleis, J.; Devreotes, P. Localization of ligand-induced phosphorylation sites to serine clusters in the c-terminal domain of the dictyostelium camp receptor, car1. J. Biol. Chem. 1994, 269, 7036–7044. [Google Scholar] [PubMed]
- Caterina, M.J.; Milne, J.L.S.; Devreotes, P.N. Mutation of the third intracellular loop of the camp receptor, car1, of dictyostelium yields mutants impaired in multiple signaling pathways. J. Biol. Chem. 1994, 269, 1523–1532. [Google Scholar] [PubMed]
- Caterina, M.J.; Hereld, D.; Devreotes, P.N. Occupancy of the dictyostelium camp receptor, car1, induces a reduction in affinity which depends upon cooh-terminal serine residues. J. Biol. Chem. 1995, 270, 4418–4423. [Google Scholar] [CrossRef] [PubMed]
- Brzostowski, J.A.; Sawai, S.; Rozov, O.; Liao, X.H.; Imoto, D.; Parent, C.A.; Kimmel, A.R. Phosphorylation of chemoattractant receptors regulates chemotaxis, actin reorganization and signal relay. J. Cell Sci. 2013, 126, 4614–4626. [Google Scholar] [CrossRef] [PubMed]
- Guetta, D.; Langou, K.; Grunwald, D.; Klein, G.; Aubry, L. Fyve-dependent endosomal targeting of an arrestin-related protein in amoeba. PLoS ONE 2010, 5, e15249. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Yan, J.; Shu, S.; Brzostowski, J.A.; Jin, T. Arrestins function in car1 gpcr-mediated signaling and car1 internalization in the development of dictyostelium discoideum. Mol. Biol. Cell 2014, 25, 3210–3221. [Google Scholar] [CrossRef] [PubMed]
- Calebiro, D.; Nikolaev, V.O.; Gagliani, M.C.; de Filippis, T.; Dees, C.; Tacchetti, C.; Persani, L.; Lohse, M.J. Persistent camp-signals triggered by internalized g-protein-coupled receptors. PLoS Biol. 2009, 7, e1000172. [Google Scholar] [CrossRef] [PubMed]
- Daaka, Y.; Luttrell, L.M.; Ahn, S.; Della Rocca, G.J.; Ferguson, S.S.; Caron, M.G.; Lefkowitz, R.J. Essential role for g protein-coupled receptor endocytosis in the activation of mitogen-activated protein kinase. J. Biol. Chem. 1998, 273, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, J.E.; Marchese, A. Regulation of gpcr trafficking by ubiquitin. Biol. Transl. Sci. 2015, 132, 15–38. [Google Scholar]
- Shenoy, S.K.; Lefkowitz, R.J. Trafficking patterns of β-arrestin and g protein-coupled receptors determined by the kinetics of β-arrestin deubiquitination. J. Biol. Chem. 2003, 278, 14498–14506. [Google Scholar] [CrossRef] [PubMed]
- Luttrell, L.M.; Gesty-Palmer, D. Beyond desensitization: Physiological relevance of arrestin-dependent signaling. Pharmacol. Rev. 2010, 62, 305–330. [Google Scholar] [CrossRef] [PubMed]
- Rosel, D.; Kimmel, A.R. The cop9 signalosome regulates cell proliferation of dictyostelium discoideum. Eur. J. Cell Biol. 2006, 85, 1023–1034. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.A.; Manning, D.R. Regulation of g proteins by covalent modification. Oncogene 2001, 20, 1643–1652. [Google Scholar] [CrossRef] [PubMed]
- Nagai, Y.; Nishimura, A.; Tago, K.; Mizuno, N.; Itoh, H. Ric-8b stabilizes the alpha subunit of stimulatory g protein by inhibiting its ubiquitination. J. Biol. Chem. 2010, 285, 11114–11120. [Google Scholar] [CrossRef] [PubMed]
- Chishiki, K.; Kamakura, S.; Yuzawa, S.; Hayase, J.; Sumimoto, H. Ubiquitination of the heterotrimeric g protein α subunits gαi2 and gαq is prevented by the guanine nucleotide exchange factor ric-8a. Biochem. Biophys. Res. Commun. 2013, 435, 414–419. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Shao, D.; Adler, M.; Charest, P.G.; Loomis, W.F.; Levine, H.; Groisman, A.; Rappel, W.J.; Firtel, R.A. Incoherent feedforward control governs adaptation of activated ras in a eukaryotic chemotaxis pathway. Sci. Signal. 2012, 5, ra2. [Google Scholar] [CrossRef] [PubMed]
- Iijima, M.; Devreotes, P. Tumor suppressor pten mediates sensing of chemoattractant gradients. Cell 2002, 109, 599–610. [Google Scholar] [CrossRef]
- Lee, S.; Comer, F.I.; Sasaki, A.; McLeod, I.X.; Duong, Y.; Okumura, K.; Yates, J.R.; Parent, C.A.; Firtel, R.A. Tor complex 2 integrates cell movement during chemotaxis and signal relay in dictyostelium. Mol. Biol. Cell 2005, 16, 4572–4583. [Google Scholar] [CrossRef] [PubMed]
- Khanna, A.; Lotfi, P.; Chavan, A.J.; Montaño, N.M.; Bolourani, P.; Weeks, G.; Shen, Z.; Briggs, S.P.; Pots, H.; Van Haastert, P.J.; et al. The small gtpases ras and rap1 bind to and control torc2 activity. Sci. Rep. 2016, 6, 25823. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Gamper, M.; Parent, C.; Firtel, R.A. The dictyostelium map kinase kinase ddmek1 regulates chemotaxis and is essential for chemoattractant-mediated activation of guanylyl cyclase. EMBO J. 1997, 16, 4317–4332. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Xu, S.; Joazeiro, C.; Cobb, M.H.; Hunter, T. The phd domain of mekk1 acts as an e3 ubiquitin ligase and mediates ubiquitination and degradation of erk1/2. Cell 2002, 9, 945–956. [Google Scholar] [CrossRef]
- Jura, N.; Bar-Sagi, D. Mapping cellular routes of ras: A ubiquitin trail. Cell Cycle 2006, 5, 2744–2747. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Park, J.; Cron, P.; Hess, D.; Hemmings, B.A. Identification of a pkb/akt hydrophobic motif ser-473 kinase as DNA-dependent protein kinase. J. Biol. Chem. 2004, 279, 41189–41196. [Google Scholar] [CrossRef] [PubMed]
- Risso, G.; Blaustein, M.; Pozzi, B.; Mammi, P.; Srebrow, A. Akt/pkb: One kinase, many modifications. Biochem. J. 2015, 468, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, J.; Gao, T. Beta-trcp-mediated ubiquitination and degradation of phlpp1 are negatively regulated by akt. Mol. Cell. Biol. 2009, 29, 6192–6205. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.L.; Wang, J.; Chan, C.H.; Lee, S.W.; Campos, A.D.; Lamothe, B.; Hur, L.; Grabiner, B.C.; Lin, X.; Darnay, B.G.; et al. The e3 ligase traf6 regulates akt ubiquitination and activation. Science 2009, 325, 1134–1138. [Google Scholar]
- Koo, J.; Wu, X.; Mao, Z.; Khuri, F.R.; Sun, S.Y. Rictor undergoes glycogen synthase kinase 3 (gsk3)-dependent, fbxw7-mediated ubiquitination and proteasomal degradation. J. Biol. Chem. 2015, 290, 14120–14129. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Wan, L.; Inuzuka, H.; Berg, A.H.; Tseng, A.; Zhai, B.; Shaik, S.; Bennett, E.; Tron, A.E.; Gasser, J.A.; et al. Rictor forms a complex with cullin-1 to promote sgk1 ubiquitination and destruction. Cell 2010, 39, 797–808. [Google Scholar] [CrossRef] [PubMed]
- Gerisch, G.; Hulser, D.; Malchow, D.; Wick, U. Cell communication by periodic cyclic-amp pulses. Philos. Trans. R. Soc. Lond. B 1975, 272, 181–192. [Google Scholar] [CrossRef]
- Pitt, G.S.; Milona, N.; Borleis, J.; Lin, K.C.; Reed, R.R.; Devreotes, P.N. Structurally distinct and stage-specific adenylyl cyclase genes play different roles in dictyostelium development. Cell 1992, 69, 305–315. [Google Scholar] [CrossRef]
- Soderbom, F.; Anjard, C.; Iranfar, N.; Fuller, D.; Loomis, W.F. An adenylyl cyclase that functions during late development of dictyostelium. Development 1999, 126, 5463–5471. [Google Scholar] [PubMed]
- Meima, M.E.; Schaap, P. Fingerprinting of adenylyl cyclase activities during dictyostelium development indicates a dominant role for adenylyl cyclase b in terminal differentiation. Dev. Biol. 1999, 212, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Majerfeld, I.H.; Leichtling, B.H.; Meligeni, J.A.; Spitz, E.; Rickenberg, H.V. A cytosolic camp dependent protein kinase in dictyostelium discoideum. I. Properties. J. Biol. Chem. 1984, 259, 654–661. [Google Scholar] [PubMed]
- De Gunzburg, J.; Veron, M. A camp-dependent protein kinase is present in differentiating dictyostelium discoideum cells. EMBO J. 1982, 1, 1063–1068. [Google Scholar]
- Mutzel, R.; Lacombe, M.L.; Simon, M.N.; de Gunzburg, J.; Veron, M. Cloning and cdna sequence of the regulatory subunit of camp-dependent protein kinase from dictyostelium discoideum. Proc. Natl. Acad. Sci. USA 1987, 84, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Kuspa, A. Dictyostelium development in the absence of camp. Science 1997, 277, 251–254. [Google Scholar] [CrossRef] [PubMed]
- Mann, S.K.O.; Firtel, R.A. A developmentally regulated, putative serine/threonine protein kinase is essential for development in dictyostelium. Mech. Dev. 1991, 35, 89–101. [Google Scholar] [CrossRef]
- Mann, S.K.O.; Brown, J.M.; Briscoe, C.; Parent, C.; Pitt, G.; Devreotes, P.N.; Firtel, R.A. Role of camp-dependent protein kinase in controlling aggregation and postaggregative development in dictyostelium. Dev. Biol. 1997, 183, 208–221. [Google Scholar] [CrossRef] [PubMed]
- Aubry, L.; Maeda, M.; Insall, R.; Devreotes, P.N.; Firtel, R.A. The dictyostelium mitogen-activated protein kinase erk2 is regulated by ras and camp-dependent protein kinase (pka) and mediates pka function. J. Biol. Chem. 1997, 272, 3883–3886. [Google Scholar] [CrossRef] [PubMed]
- Laub, M.T.; Loomis, W.F. A molecular network that produces spontaneous oscillations in excitable cells of dictyostelium. Mol. Biol. Cell 1998, 9, 3521–3532. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.N.; Pelegrini, O.; Veron, M.; Kay, R.R. Mutation of protein kinase a causes heterochronic development of dictyostelium. Nature 1992, 356, 171–172. [Google Scholar] [CrossRef] [PubMed]
- Shaulsky, G.; Fuller, D.; Loomis, W.F. A camp-phosphodiesterase controls pka-dependent differentiation. Development 1998, 125, 691–699. [Google Scholar] [PubMed]
- Thomason, P.A.; Traynor, D.; Cavet, G.; Chang, W.T.; Harwood, A.J.; Kay, R.R. An intersection of the camp/pka and two-component signal transduction systems in dictyostelium. EMBO J. 1998, 17, 2838–2845. [Google Scholar] [CrossRef] [PubMed]
- Michel, J.J.; Scott, J.D. Akap mediated signal transduction. Annu. Rev. Pharmacol. Toxicol. 2002, 42, 235–257. [Google Scholar] [CrossRef] [PubMed]
- Welch, E.J.; Jones, B.W.; Scott, J.D. Networking with akaps: Context-dependent regulation of anchored enzymes. Mol. Interv. 2010, 10, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Dell’Acqua, M.L.; Smith, K.E.; Gorski, J.A.; Horne, E.A.; Gibson, E.S.; Gomez, L.L. Regulation of neuronal pka signaling through akap targeting dynamics. Eur. J. Cell Biol. 2006, 85, 627–633. [Google Scholar] [CrossRef] [PubMed]
- Lignitto, L.; Carlucci, A.; Sepe, M.; Stefan, E.; Cuomo, O.; Nistico, R. Control of pka stability and signalling by the ring ligase praja2. Nat. Cell Biol. 2011, 13, 412–422. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Katoh-Kurasawa, M.; Muramoto, T.; Santhanam, B.; Long, Y.; Li, L.; Ueda, M.; Iglesias, P.A.; Shaulsky, G.; Devreotes, P.N. Nucleocytoplasmic shuttling of a gata transcription factor functions as a development timer. Science 2014, 343, 1249531. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, T.; Kitagawa, K.; Ohhata, T.; Sakai, S.; Uchida, C.; Shibata, K.; Minegishi, N.; Yumimoto, K.; Nakayama, K.I.; Masumoto, K.; et al. Regulation of gata-binding protein 2 levels via ubiquitin-dependent degradation by fbw7:Involvement of cyclin b-cyclin-dependent kinase 1-mediated phosphorylation of thr176 in gata-binding protein 2. J. Biol. Chem. 2015, 290, 10368–10381. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, K.; Shibata, K.; Matsumoto, A.; Matsumoto, M.; Ohhata, T.; Nakayama, K.I.; Niida, H.; Kitagawa, M. Fbw7 targets gata3 through cyclin-dependent kinase 2-dependent proteolysis and contributes to regulation of t-cell development. Mol. Cell. Biol. 2014, 34, 2732–2744. [Google Scholar] [CrossRef] [PubMed]
- Pergolizzi, B.; Peracino, B.; Silverman, J.; Ceccarelli, A.; Noegel, A.; Devreotes, P.; Bozzaro, S. Temperature-sensitive inhibition of development in dictyostelium due to a point mutation in the piaa gene. Dev. Biol. 2002, 251, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Tena, S.; Cubillos-Rojas, M.; Schneider, T.; Rosa, J. Functional and pathological relevance of herc family proteins: A decade later. Cell Mol. Life Sci. 2016, 73, 1955–1968. [Google Scholar] [CrossRef] [PubMed]
- Rotin, D.; Kumar, S. Physiological functions of the hect family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2009, 10, 398–409. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Zhang, N.; Murphy, D.B.; Devreotes, P.N. Dynamic distribution of chemoattractant receptors in living cells during chemotaxis and persistent stimulation. J. Cell Biol. 1997, 139, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Serge, A.; de Keijzer, S.; Van Hemert, F.; Hickman, M.R.; Hereld, D.; Spaink, H.P.; Schmidt, T.; Snaar-Jagalska, B.E. Quantification of gpcr internalization by single-molecule microscopy in living cells. Integr. Biol. (Camb.) 2011, 3, 675–683. [Google Scholar] [CrossRef] [PubMed]
- Chan, N.C.; den Besten, W.; Sweredoski, M.J.; Hess, S.; Deshaies, R.J.; Chan, D.C. Degradation of the deubiquitinating enzyme usp33 is mediated by p97 and the ubiquitin ligase herc2. J. Biol. Chem. 2014, 289, 19789–19798. [Google Scholar] [CrossRef] [PubMed]




© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pergolizzi, B.; Bozzaro, S.; Bracco, E. G-Protein Dependent Signal Transduction and Ubiquitination in Dictyostelium. Int. J. Mol. Sci. 2017, 18, 2180. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18102180
Pergolizzi B, Bozzaro S, Bracco E. G-Protein Dependent Signal Transduction and Ubiquitination in Dictyostelium. International Journal of Molecular Sciences. 2017; 18(10):2180. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18102180
Chicago/Turabian StylePergolizzi, Barbara, Salvatore Bozzaro, and Enrico Bracco. 2017. "G-Protein Dependent Signal Transduction and Ubiquitination in Dictyostelium" International Journal of Molecular Sciences 18, no. 10: 2180. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18102180