Proteases of Dermatophagoides pteronyssinus
Abstract
:1. Introduction
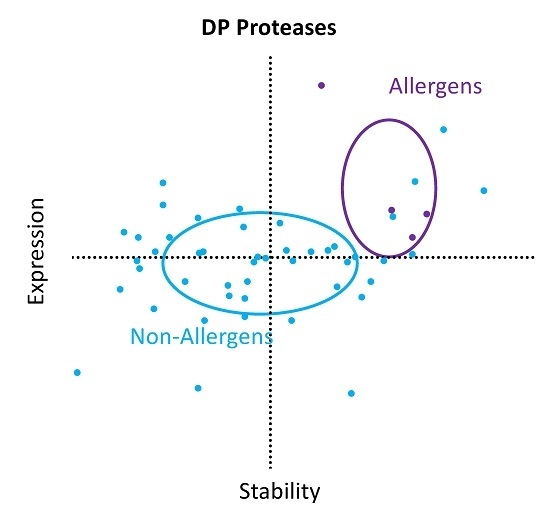
2. Results
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| DP | Dermatophagoides pteronyssinus |
| DF | Dermatophagoides farinae |
| SS | Sarcoptes scabiei |
| GND½ | Concentration of Guanidinium Chloride at the Transition Midpoint of the Chemical Denaturation Curve |
| SPROX | Stability of Proteins from Rates of Oxidation |
| FPKM | Fragments per Kilobase per Million Reads |
References
- Puente, X.S.; Sanchez, L.M.; Overall, C.M.; Lopez-Otin, C. Human and mouse proteases: A comparative genomic approach. Nat. Rev. Genet. 2003, 4, 544–558. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, N.D.; Barrett, A.J.; Finn, R. Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2016, 44, D343–D350. [Google Scholar] [CrossRef] [PubMed]
- Radauer, C.; Breiteneder, H. Pollen allergens are restricted to few protein families and show distinct patterns of species distribution. J. Allergy Clin. Immunol. 2006, 117, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Tovey, E.R.; Chapman, M.D.; Platts-Mills, T.A. Mite faeces are a major source of house dust allergens. Nature 1981, 289, 592–593. [Google Scholar] [CrossRef] [PubMed]
- Chua, K.Y.; Stewart, G.A.; Thomas, W.R.; Simpson, R.J.; Dilworth, R.J.; Plozza, T.M.; Turner, K.J. Sequence analysis of cDNA coding for a major house dust mite allergen, Der p 1. Homology with cysteine proteases. J. Exp. Med. 1988, 167, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.; Farmer, K.; MacDonald, L.; Kalsheker, N.; Pritchard, D.; Haslett, C.; Lamb, J.; Sallenave, J.M. House dust mite Der p 1 downregulates defenses of the lung by inactivating elastase inhibitors. Am. J. Respir. Cell Mol. 2003, 29, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Deb, R.; Shakib, F.; Reid, K.; Clark, H. Major house dust mite allergens Dermatophagoides pteronyssinus 1 and Dermatophagoides farinae 1 degrade and inactivate lung surfactant proteins A and D. J. Biol. Chem. 2007, 282, 36808–36819. [Google Scholar] [CrossRef] [PubMed]
- Lewkowich, I.P.; Day, S.B.; Ledford, J.R.; Zhou, P.; Dienger, K.; Wills-Karp, M.; Page, K. Protease-activated receptor 2 activation of myeloid dendritic cells regulates allergic airway inflammation. Respir. Res. 2011, 12, 122. [Google Scholar] [CrossRef] [PubMed]
- Gough, L.; Schulz, O.; Sewell, H.F.; Shakib, F. The cysteine protease activity of the major dust mite allergen Der p 1 selectively enhances the immunoglobulin E antibody response. J. Exp. Med. 1999, 190, 1897–1901. [Google Scholar] [CrossRef] [PubMed]
- Ghaemmaghami, A.M.; Shakib, F. Human t cells that have been conditioned by the proteolytic activity of the major dust mite allergen Der p 1 trigger enhanced immunoglobulin E synthesis by B cells. Clin. Exp. Allergy 2002, 32, 728–732. [Google Scholar] [CrossRef] [PubMed]
- Chapman, M.D.; Wunschmann, S.; Pomes, A. Proteases as Th2 adjuvants. Curr. Allergy Asthma Rep. 2007, 7, 363–367. [Google Scholar] [CrossRef] [PubMed]
- Morales, M.; Iraola, V.; Leonor, J.R.; Carnes, J. Enzymatic activity of allergenic house dust and storage mite extracts. J. Med. Entomol. 2013, 50, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Vidal-Quist, J.C.; Ortego, F.; Castanera, P.; Hernandez-Crespo, P. Quality control of house dust mite extracts by broad-spectrum profiling of allergen-related enzymatic activities. Allergy 2017, 72, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Sudha, V.T.; Arora, N.; Gaur, S.N.; Pasha, S.; Singh, B.P. Identification of a serine protease as a major allergen (Per a 10) of Periplaneta americana. Allergy 2008, 63, 768–776. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.D.; Tam, M.F.; Chou, H.; Han, S.H. The importance of serine proteinases as aeroallergens associated with asthma. Int. Arch. Allergy Immunol. 1999, 119, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Bouley, J.; Groeme, R.; Le Mignon, M.; Jain, K.; Chabre, H.; Bordas-Le Floch, V.; Couret, M.N.; Bussieres, L.; Lautrette, A.; Naveau, M.; et al. Identification of the cysteine protease Amb a 11 as a novel major allergen from short ragweed. J. Allergy Clin. Immunol. 2015, 136, 1055–1064. [Google Scholar] [CrossRef] [PubMed]
- Pastorello, E.A.; Conti, A.; Pravettoni, V.; Farioli, L.; Rivolta, F.; Ansaloni, R.; Ispano, M.; Incorvaia, C.; Giuffrida, M.G.; Ortolani, C. Identification of actinidin as the major allergen of kiwi fruit. J. Allergy Clin. Immunol. 1998, 101, 531–537. [Google Scholar] [CrossRef]
- Yasueda, H.; Mita, H.; Akiyama, K.; Shida, T.; Ando, T.; Sugiyama, S.; Yamakawa, H. Allergens from Dermatophagoides mites with chymotryptic activity. Clin. Exp. Allergy 1993, 23, 384–390. [Google Scholar] [CrossRef] [PubMed]
- King, C.; Simpson, R.J.; Moritz, R.L.; Reed, G.E.; Thompson, P.J.; Stewart, G.A. The isolation and characterization of a novel collagenolytic serine protease allergen (Der p 9) from the dust mite Dermatophagoides pteronyssinus. J. Allergy Clin. Immunol. 1996, 98, 739–747. [Google Scholar] [CrossRef]
- Thomas, W.R. Hierarchy and molecular properties of house dust mite allergens. Allergol. Int. 2015, 64, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Resch, Y.; Michel, S.; Kabesch, M.; Lupinek, C.; Valenta, R.; Vrtala, S. Different IgE recognition of mite allergen components in asthmatic and nonasthmatic children. J. Allergy Clin. Immunol. 2015, 136, 1083–1091. [Google Scholar] [CrossRef] [PubMed]
- Erban, T.; Harant, K.; Hubert, J. Detailed two-dimensional gel proteomic mapping of the feces of the house dust mite Dermatophagoides pteronyssinus and comparison with D. farinae: Reduced trypsin protease content in D. pteronyssinus and different isoforms. J. Proteom. 2017. [Google Scholar] [CrossRef] [PubMed]
- Batard, T.; Baron-Bodo, V.; Martelet, A.; Le Mignon, M.; Lemoine, P.; Jain, K.; Mariano, S.; Horiot, S.; Chabre, H.; Harwanegg, C.; et al. Patterns of IgE sensitization in house dust mite-allergic patients: Implications for allergen immunotherapy. Allergy 2016, 71, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Mueller, G.A.; Randall, T.A.; Glesner, J.; Pedersen, L.C.; Perera, L.; Edwards, L.L.; DeRose, E.F.; Chapman, M.D.; London, R.E.; Pomes, A. Serological, genomic and structural analyses of the major mite allergen Der p 23. Clin. Exp. Allergy 2016, 46, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Ogburn, R.N.; Randall, T.A.; Xu, Y.; Roberts, J.H.; Mebrahtu, B.; Karnuta, J.M.; Rider, S.D.; Kissling, G.E.; London, R.E.; Pomes, A.; et al. Are dust mite allergens more abundant and/or more stable than other Dermatophagoides pteronyssinus proteins? J. Allergy Clin. Immunol. 2017, 139, 1030–1032. [Google Scholar] [CrossRef] [PubMed]
- Randall, T.A.; Mullikin, J. NISC Comparative Sequencing Program; Mueller, G.A. A draft genome assembly and analysis of the house dust mite Dermatophagoides pteronyssinus. Allergy 2017. submitted. [Google Scholar]
- Chan, T.F.; Ji, K.M.; Yim, A.K.; Liu, X.Y.; Zhou, J.W.; Li, R.Q.; Yang, K.Y.; Li, J.; Li, M.; Law, P.T.; et al. The draft genome, transcriptome, and microbiome of dermatophagoides farinae reveal a broad spectrum of dust mite allergens. J. Allergy Clin. Immunol. 2015, 135, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Rider, S.D.; Morgan, M.S.; Arlian, L.G. Draft genome of the scabies mite. Parasite Vector 2015, 8, 585. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C.; Shen, H.B. Protident: A web server for identifying proteases and their types by fusing functional domain and sequential evolution information. Biochem. Biophys. Res. Commun. 2008, 376, 321–325. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Coleman-Derr, D.; Chen, G.P.; Gu, Y.Q. OrthoVenn: A web server for genome wide comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2015, 43, W78–W84. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Strickland, E.C.; Fitzgerald, M.C. Thermodynamic analysis of protein folding and stability using a tryptophan modification protocol. Anal. Chem. 2014, 86, 7041–7048. [Google Scholar] [CrossRef] [PubMed]
- West, G.M.; Tang, L.; Fitzgerald, M.C. Thermodynamic analysis of protein stability and ligand binding using a chemical modification- and mass-spectrometry based strategy. Anal. Chem. 2008, 80, 4175–4185. [Google Scholar] [CrossRef] [PubMed]
- Manly, B.F.J. Multivariate Statistical Methods a Primer; Chapman and Hall: London, UK, 1986. [Google Scholar]
- Vogel, C.; Marcotte, E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012, 13, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Avula-Poola, S.; Morgan, M.S.; Arlian, L.G. Diet influences growth rates and allergen and endotoxin contents of cultured Dermatophagoides farinae and Dermatophagoides pteronyssinus house dust mites. Int. Arch. Allergy Immunol. 2012, 159, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Thomas, W.R. House dust mite allergens: New discoveries and relevance to the allergic patient. Curr. Allergy Asthma Rep. 2016, 16, 69. [Google Scholar] [CrossRef] [PubMed]
- Bonnelykke, K.; Sparks, R.; Waage, J.; Milner, J.D. Genetics of allergy and allergic sensitization: Common variants, rare mutations. Curr. Opin. Immunol. 2015, 36, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Platts-Mills, T.A.E. The allergy epidemics: 1870–2010. J. Allergy Clin. Immunol. 2015, 136, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Gilles-Stein, S.; Traidl-Hoffmann, C. Pollen are more than allergen carriers. Allergologie 2016, 39, 69–76. [Google Scholar] [CrossRef]
- Baldacci, S.; Maio, S.; Cerrai, S.; Sarno, G.; Baiz, N.; Simoni, M.; Annesi-Maesano, I.; Viegi, G.; Study, H. Allergy and asthma: Effects of the exposure to particulate matter and biological allergens. Respir. Med. 2015, 109, 1089–1104. [Google Scholar] [CrossRef] [PubMed]
- Arlian, L.G.; Morgan, M.S. Immunomodulation of skin cytokine secretion by house dust mite extracts. Int. Arch. Allergy Immunol. 2011, 156, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Arlian, L.G.; Morgan, M.S.; Rider, S.D. Sarcoptes scabiei: Genomics to proteomics to biology. Parasite Vector 2016, 9, 380. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. Accelerated profile hmm searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. Mafft multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]





| Protease Type | D. pteronyssinus | D. farinae | S. scabiei |
|---|---|---|---|
| Threonine Proteases | 18 | 20 | 17 |
| Serine Proteases | 133 | 88 | 76 |
| Metalloproteases | 127 | 94 | 83 |
| Cysteine Proteases | 77 | 60 | 61 |
| Aspartyl Proteases | 14 | 5 | 6 |
| Total | 369 | 267 | 243 |
| Data Compared | Group 1 | n1 | Group 2 | n2 | p Value | Corrected p † |
|---|---|---|---|---|---|---|
| log10 FPKM | ||||||
| all mite | 14,406 | proteases | 255 | <0.001 | ||
| asp | 11 | cys | 61 | 0.288 | 3.170 | |
| asp | 11 | metallo | 83 | 0.204 | 2.243 | |
| asp | 11 | ser | 84 | 0.172 | 1.891 | |
| asp | 11 | thr | 16 | 0.003 | 0.033 | |
| cys | 61 | metallo | 83 | 0.747 | 8.220 | |
| cys | 61 | ser | 84 | 0.783 | 8.612 | |
| cys | 61 | thr | 16 | 0.052 | 0.576 | |
| metallo | 83 | ser | 84 | 0.943 | 10.375 | |
| metallo | 83 | thr | 16 | 0.004 | 0.040 | |
| ser | 84 | thr | 16 | 0.003 | 0.029 | |
| t Tests FPKM and GND½ * | ||||||
| GND½ | ||||||
| cys | 9 | metallo | 18 | 0.005 | 0.035 | |
| cys | 9 | ser | 12 | 0.043 | 0.301 | |
| cys | 9 | thr | 10 | 0.060 | 0.420 | |
| metallo | 18 | ser | 12 | 0.260 | 1.820 | |
| metallo | 18 | thr | 10 | 0.135 | 0.945 | |
| ser | 12 | thr | 10 | 0.178 | 1.246 | |
| log10 FPKM | ||||||
| allergens | 4 | non-allergens | 45 | 0.010 | 0.070 | |
| cys | 9 | metallo | 18 | 0.490 | 3.430 | |
| cys | 9 | ser | 12 | 0.851 | 5.957 | |
| cys | 9 | thr | 10 | 0.092 | 0.644 | |
| metallo | 18 | ser | 12 | 0.266 | 1.862 | |
| metallo | 18 | thr | 10 | 0.135 | 0.945 | |
| ser | 12 | thr | 10 | 0.018 | 0.126 | |
| allergens | 4 | non-allergens | 45 | 0.007 | 0.049 |
| Data 1 | Data 2 | Group 1 | Group 2 | T2 | F | df1 | df2 | p | Corrected p † |
|---|---|---|---|---|---|---|---|---|---|
| GND½ * | log10 FPKM | cys | metallo | 9.9 | 4.8 | 2 | 24 | 0.013 | 0.091 |
| cys | ser | 6.9 | 3.2 | 2 | 18 | 0.047 | 0.329 | ||
| cys | thr | 5.5 | 2.6 | 2 | 16 | 0.081 | 0.567 | ||
| metallo | ser | 2.6 | 1.3 | 2 | 27 | 0.300 | 2.100 | ||
| metallo | thr | 5.6 | 2.7 | 2 | 25 | 0.072 | 0.504 | ||
| ser | thr | 6.8 | 3.2 | 2 | 19 | 0.046 | 0.322 | ||
| allergens | non-allergens | 11.9 | 5.8 | 2 | 46 | 0.004 | 0.028 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Randall, T.A.; London, R.E.; Fitzgerald, M.C.; Mueller, G.A. Proteases of Dermatophagoides pteronyssinus. Int. J. Mol. Sci. 2017, 18, 1204. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18061204
Randall TA, London RE, Fitzgerald MC, Mueller GA. Proteases of Dermatophagoides pteronyssinus. International Journal of Molecular Sciences. 2017; 18(6):1204. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18061204
Chicago/Turabian StyleRandall, Thomas A., Robert E. London, Michael C. Fitzgerald, and Geoffrey A. Mueller. 2017. "Proteases of Dermatophagoides pteronyssinus" International Journal of Molecular Sciences 18, no. 6: 1204. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18061204