The Role of PPARβ/δ in Melanoma Metastasis
Abstract
:1. Introduction
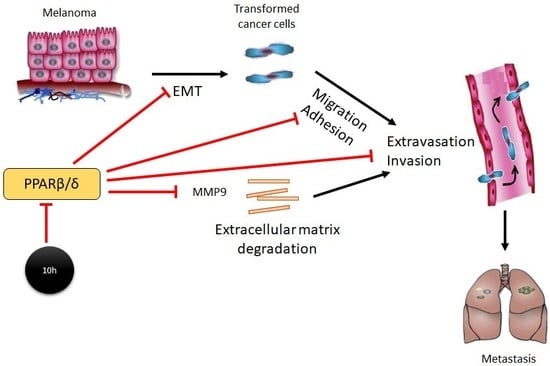
2. Results
2.1. 10h Induces Phenotypic Changes of B16/F10 Mouse Melanoma Cells
2.2. 10h Promotes Melanoma Cell Migration and Invasion
2.3. 10h Increases B16/F10 Melanoma Cell Adhesion to the Endothelium and Trans-Endothelial Migration
2.4. PPARβ/δ Inhibition Promotes Lung Metastasis of Melanoma Cells In Vivo
3. Discussion
4. Material and Methodology
4.1. Animals
4.2. Cell Culture
4.3. Synthesis of 10h
4.4. RNA Extraction and Quantitative Real-time PCR
4.5. Epithelial Mesenchymal Transition (EMT)
4.6. Measurement of Extracellular Melanin Content
4.7. Cell Adhesion and Tumour-Endothelial Assay
4.8. Trans-Well Migration and Invasion Assay
4.9. Trans-Endothelial Migration
4.10. In Vivo Metastasis Assay
4.11. Extravasation Assay
4.12. Evans Blue Assay.
4.13. Statistics
Author Contributions
Funding
Conflicts of Interest
References
- Apalla, Z.; Lallas, A.; Sotiriou, E.; Lazaridou, E.; Ioannides, D. Epidemiological trends in skin cancer. Dermatol. Pract. Concept. 2017, 7, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Tas, F. Metastatic behavior in melanoma: Timing, pattern, survival, and influencing factors. J. Oncol. 2012, 2012, 647684. [Google Scholar] [CrossRef] [PubMed]
- Pawlik, T.M.; Sondak, V.K. Malignant melanoma: Current state of primary and adjuvant treatment. Crit. Rev. Oncol. Hematol. 2003, 45, 245–264. [Google Scholar] [CrossRef]
- Balch, C.M.; Gershenwald, J.E.; Soong, S.J.; Thompson, J.F.; Atkins, M.B.; Byrd, D.R.; Buzaid, A.C.; Cochran, A.J.; Coit, D.G.; Ding, S.; et al. Final version of 2009 AJCC melanoma staging and classification. J. Clin. Oncol. 2009, 27, 6199–6206. [Google Scholar] [CrossRef] [PubMed]
- Sandru, A.; Voinea, S.; Panaitescu, E.; Blidaru, A. Survival rates of patients with metastatic malignant melanoma. J. Med. Life 2014, 7, 572–576. [Google Scholar] [PubMed]
- Magadum, A.; Engel, F. PPARβ/δ: Linking metabolism to regeneration. Int. J. Mol. Sci. 2018, 19, 2013. [Google Scholar] [CrossRef] [PubMed]
- Dreyer, C.; Krey, G.; Keller, H.; Givel, F.; Helftenbein, G.; Wahli, W. Control of the peroxisomal beta-oxidation pathway by a novel family of nuclear hormone receptors. Cell 1992, 68, 879–887. [Google Scholar] [CrossRef]
- Wang, X.; Wang, G.; Shi, Y.; Sun, L.; Gorczynski, R.; Li, Y.J.; Xu, Z.; Spaner, D.E. Ppar-delta promotes survival of breast cancer cells in harsh metabolic conditions. Oncogenesis 2016, 5, e232. [Google Scholar] [CrossRef] [PubMed]
- Zuo, X.; Xu, W.; Xu, M.; Tian, R.; Moussalli, M.J.; Mao, F.; Zheng, X.; Wang, J.; Morris, J.S.; Gagea, M.; et al. Metastasis regulation by PPARD expression in cancer cells. JCI Insight 2017, 2, e91419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muller-Brusselbach, S.; Komhoff, M.; Rieck, M.; Meissner, W.; Kaddatz, K.; Adamkiewicz, J.; Keil, B.; Klose, K.J.; Moll, R.; Burdick, A.D.; et al. Deregulation of tumor angiogenesis and blockade of tumor growth in PPARbeta-deficient mice. EMBO J. 2007, 26, 3686–3698. [Google Scholar] [CrossRef] [PubMed]
- Montagner, A.; Delgado, M.B.; Tallichet-Blanc, C.; Chan, J.S.K.; Sng, M.K.; Mottaz, H.; Degueurce, G.; Lippi, Y.; Moret, C.; Baruchet, M.; et al. Src is activated by the nuclear receptor peroxisome proliferator-activated receptor beta/delta in ultraviolet radiation-induced skin cancer. EMBO Mol. Med. 2014, 6, 80–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, H.Y.; Chung, E.; Lee, M.; Cho, Y.; Kang, W.H. Expression and function of peroxisome proliferator-activated receptors in human melanocytes. Br. J. Dermatol. 2004, 150, 462–468. [Google Scholar] [CrossRef] [PubMed]
- Borland, M.G.; Yao, P.L.; Kehres, E.M.; Lee, C.; Pritzlaff, A.M.; Ola, E.; Wagner, A.L.; Shannon, B.E.; Albrecht, P.P.; Zhu, B.K.; et al. Editor’s highlight: PPAR beta/delta and PPAR gamma inhibit melanoma tumorigenicity by modulating inflammation and apoptosis. Toxicol. Sci. 2017, 159, 436–448. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.E.; Leung, E.; Baguley, B.C.; Finlay, G.J. Heterogeneity of expression of epithelial-mesenchymal transition markers in melanocytes and melanoma cell lines. Front. Genet. 2013, 4, 97. [Google Scholar] [CrossRef] [PubMed]
- Toth, P.M.; Naruhn, S.; Pape, V.F.S.; Dorr, S.M.A.; Klebe, G.; Muller, R.; Diederich, W.E. Development of improved PPAR ss/d inhibitors. Chemmedchem 2012, 7, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Sng, M.K.; Chan, J.S.K.; Teo, Z.; Phua, T.; Tan, E.H.P.; Wee, J.W.K.; Koh, N.J.N.; Tan, C.K.; Chen, J.P.; Pal, M.; et al. Selective deletion of PPARbeta/delta in fibroblasts causes dermal fibrosis by attenuated LRG1 expression. Cell Discov. 2018, 4, 15. [Google Scholar] [CrossRef] [PubMed]
- Galaup, A.; Cazes, A.; Le Jan, S.; Philippe, J.; Connault, E.; Le Coz, E.; Mekid, H.; Mir, L.M.; Opolon, P.; Corvol, P.; et al. Angiopoietin-like 4 prevents metastasis through inhibition of vascular permeability and tumor cell motility and invasiveness. Proc. Natl. Acad. Sci. USA 2006, 103, 18721–18726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carreira, S.; Goodall, J.; Denat, L.; Rodriguez, M.; Nuciforo, P.; Hoek, K.S.; Testori, A.; Larue, L.; Goding, C.R. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev. 2006, 20, 3426–3439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Mello, S.A.; Finlay, G.J.; Baguley, B.C.; Askarian-Amiri, M.E. Signaling pathways in melanogenesis. Int. J. Mol. Sci. 2016, 17, 1144. [Google Scholar] [CrossRef] [PubMed]
- Wendt, M.K.; Allington, T.M.; Schiemann, W.P. Mechanisms of the epithelial-mesenchymal transition by TGF-beta. Future Oncol. 2009, 5, 1145–1168. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.M.; Bissell, M.J. Of extracellular matrix, scaffolds, and signaling: Tissue architecture regulates development, homeostasis, and cancer. Annu. Rev. Cell Dev. Biol. 2006, 22, 287–309. [Google Scholar] [CrossRef] [PubMed]
- Gaggioli, C.; Hooper, S.; Hidalgo-Carcedo, C.; Grosse, R.; Marshall, J.F.; Harrington, K.; Sahai, E. Fibroblast-led collective invasion of carcinoma cells with differing roles for RHOGTPases in leading and following cells. Nat. Cell Biol. 2007, 9, 1392–1400. [Google Scholar] [CrossRef] [PubMed]
- Kessenbrock, K.; Plaks, V.; Werb, Z. Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell 2010, 141, 52–67. [Google Scholar] [CrossRef] [PubMed]
- Gialeli, C.; Theocharis, A.D.; Karamanos, N.K. Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J. 2011, 278, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Radisky, E.S.; Radisky, D.C. Matrix metalloproteinase-induced epithelial-mesenchymal transition in breast cancer. J. Mammary Gland. Biol. Neoplasia 2010, 15, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Orgaz, J.L.; Pandya, P.; Dalmeida, R.; Karagiannis, P.; Sanchez-Laorden, B.; Viros, A.; Albrengues, J.; Nestle, F.O.; Ridley, A.J.; Gaggioli, C.; et al. Diverse matrix metalloproteinase functions regulate cancer amoeboid migration. Nat. Commun. 2014, 5, 4255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, W.H.; Liu, H.; Xu, S.; Entschladen, F.; Niggemann, B.; Zänker, K.S.; Han, R. Migration and metalloproteinases determine the invasive potential of mouse melanoma cells, but not melanin and telomerase. Cancer Lett. 2001, 162, S49–S55. [Google Scholar] [CrossRef]
- Shellman, Y.G.; Makela, M.; Norris, D.A. Induction of secreted matrix metalloproteinase-9 activity in human melanoma cells by extracellular matrix proteins and cytokines. Melanoma Res. 2006, 16, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Cavallaro, U.; Christofori, G. Cell adhesion in tumor invasion and metastasis: Loss of the glue is not enough. Biochim. Biophys. Acta 2001, 1552, 39–45. [Google Scholar] [CrossRef]
- Onken, M.D.; Mooren, O.L.; Mukherjee, S.; Shahan, S.T.; Li, J.; Cooper, J.A. Endothelial monolayers and transendothelial migration depend on mechanical properties of the substrate. Cytoskeleton 2014, 71, 695–706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ham, S.A.; Yoo, T.; Hwang, J.S.; Kang, E.S.; Lee, W.J.; Paek, K.S.; Park, C.; Kim, J.H.; Do, J.T.; Lim, D.S.; et al. Ligand-activated PPARδ modulates the migration and invasion of melanoma cells by regulating Snail expression. Am. J. Cancer Res. 2014, 4, 674. [Google Scholar] [PubMed]
- Berger, J.; Moller, D.E. The mechanisms of action of PPARs. Annu. Rev. Med. 2002, 53, 409–435. [Google Scholar] [CrossRef] [PubMed]
- Feige, J.N.; Gelman, L.; Michalik, L.; Desvergne, B.; Wahli, W. From molecular action to physiological outputs: Peroxisome proliferator-activated receptors are nuclear receptors at the crossroads of key cellular functions. Prog. Lipid Res. 2006, 45, 120–159. [Google Scholar] [CrossRef] [PubMed]
- Youssef, J.; Badr, M. Peroxisome proliferator-activated receptors and cancer: Challenges and opportunities. Br. J. Pharmacol. 2011, 164, 68–82. [Google Scholar] [CrossRef] [PubMed]
- Muller, R. PPARbeta/delta in human cancer. Biochimie 2017, 136, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Michalik, L.; Wahli, W. PPARs mediate lipid signaling in inflammation and cancer. PPAR Res. 2008, 2008, 134059. [Google Scholar] [CrossRef] [PubMed]
- Coleman, J.D.; Thompson, J.T.; Smith, R.W., 3rd.; Prokopczyk, B.; Vanden Heuvel, J.P. Role of peroxisome proliferator-activated receptor beta/delta and B-cell lymphoma-6 in regulation of genes involved in metastasis and migration in pancreatic cancer cells. PPAR Res. 2013, 2013, 121956. [Google Scholar] [CrossRef] [PubMed]
- Falzone, L.R.; Travali, S.; Scalisi, A.; McCubrey, J.A.; Candido, S. Libra M1. MMP-9 overexpression is associated with intragenic hypermethylation of MMP9 gene in melanoma. Aging 2016, 8, 933. [Google Scholar] [CrossRef] [PubMed]
- Heerboth, S.; Housman, G.; Leary, M.; Longacre, M.; Byler, S.; Lapinska, K.; Willbanks, A.; Sarkar, S. EMT and tumor metastasis. Clin. Transl. Med. 2015, 4, 6. [Google Scholar] [CrossRef] [PubMed]
- Steingrimsson, E.; Copeland, N.G.; Jenkins, N.A. Melanocytes and the microphthalmia transcription factor network. Annu. Rev Genet. 2004, 38, 365–411. [Google Scholar] [CrossRef] [PubMed]
- Vance, K.W.; Goding, C.R. The transcription network regulating melanocyte development and melanoma. Pigment Cell Res. 2004, 17, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Pinner, S.; Jordan, P.; Sharrock, K.; Bazley, L.; Collinson, L.; Marais, R.; Bonvin, E.; Goding, C.; Sahai, E. Intravital imaging reveals transient changes in pigment production and Brn2 expression during metastatic melanoma dissemination. Cancer Res. 2009, 69, 7969–7977. [Google Scholar] [CrossRef] [PubMed]
- Thurber, A.E.; Douglas, G.; Sturm, E.C.; Zabierowski, S.E.; Smit, D.J.; Ramakrishnan, S.N.; Hacker, E.; Leonard, J.H.; Herlyn, M.; Sturm, R.A. Inverse expression states of the BRN2 and MITF transcription factors in melanoma spheres and tumour xenografts regulate the NOTCH pathway. Oncogene 2011, 30, 3036–3048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bell, R.E.; Khaled, M.; Netanely, D.; Schubert, S.; Golan, T.; Buxbaum, A.; Janas, M.M.; Postolsky, B.; Goldberg, M.S.; Shamir, R.; et al. Transcription factor/microRNA axis blocks melanoma invasion program by miR-211 targeting NUAK1. J. Investig. Dermatol. 2014, 134, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Cheli, Y.; Giuliano, S.; Fenouille, N.; Allegra, M.; Hofman, V.; Hofman, P.; Bahadoran, P.; Lacour, J.P.; Tartare-Deckert, S.; Bertolotto, C.; et al. Hypoxia and MITF control metastatic behaviour in mouse and human melanoma cells. Oncogene 2012, 31, 2461–2470. [Google Scholar] [CrossRef] [PubMed]
- Ombrato, L.; Malanchi, I. The EMT universe: Space between cancer cell dissemination and metastasis initiation. Crit. Rev. Oncog. 2014, 19, 349–361. [Google Scholar] [PubMed]
- Tan, N.S.; Vazquez-Carrera, M.; Montagner, A.; Sng, M.K.; Guillou, H.; Wahli, W. Transcriptional control of physiological and pathological processes by the nuclear receptor PPAR beta/delta. Prog. Lipid Res. 2016, 64, 98–122. [Google Scholar] [CrossRef] [PubMed]
- Nadra, K.; Anghel, S.I.; Joye, E.; Tan, N.S.; Basu-Modak, S.; Trono, D.; Wahli, W.; Desvergne, B. Differentiation of trophoblast giant cells and their metabolic functions are dependent on peroxisome proliferator-activated receptor beta/delta. Mol. Cell. Boil. 2006, 26, 3266–3281. [Google Scholar]




| Target (Mouse) | Forward Sequence (5′–3′) | Reverse Sequence (5′–3′) |
|---|---|---|
| Angptl4 (NM_020581.2) | GGGACCTTAACTGTGCCAAGAG | GGAAGTATTGTCCATTGAGATTGGA |
| Cpt1a (NM_013495.2) | ACACCATCCACGCCATACTG | TCCCAGAAGACGAATAGGTTTGAG |
| Mmp9 (NM_013599.4) | GCCCTGGAACTCACACGACA | TTGGAAACTCACACGCCAGAAG |
| Gapdh (NM_001289726.1) | ACTGAGGACCAGGTTGTCTCC | CTGTAGCCGTATTCATTGTCATACC |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lim, J.C.W.; Kwan, Y.P.; Tan, M.S.; Teo, M.H.Y.; Chiba, S.; Wahli, W.; Wang, X. The Role of PPARβ/δ in Melanoma Metastasis. Int. J. Mol. Sci. 2018, 19, 2860. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19102860
Lim JCW, Kwan YP, Tan MS, Teo MHY, Chiba S, Wahli W, Wang X. The Role of PPARβ/δ in Melanoma Metastasis. International Journal of Molecular Sciences. 2018; 19(10):2860. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19102860
Chicago/Turabian StyleLim, Jonathan Chee Woei, Yuet Ping Kwan, Michelle Siying Tan, Melissa Hui Yen Teo, Shunsuke Chiba, Walter Wahli, and Xiaomeng Wang. 2018. "The Role of PPARβ/δ in Melanoma Metastasis" International Journal of Molecular Sciences 19, no. 10: 2860. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19102860