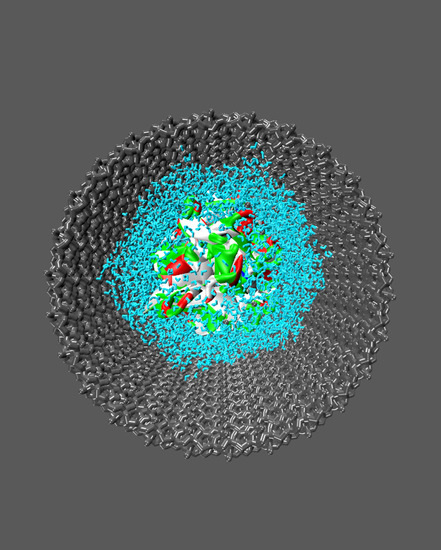
Confining a Protein-Containing Water Nanodroplet inside Silica Nanochannels
Abstract
:1. Introduction
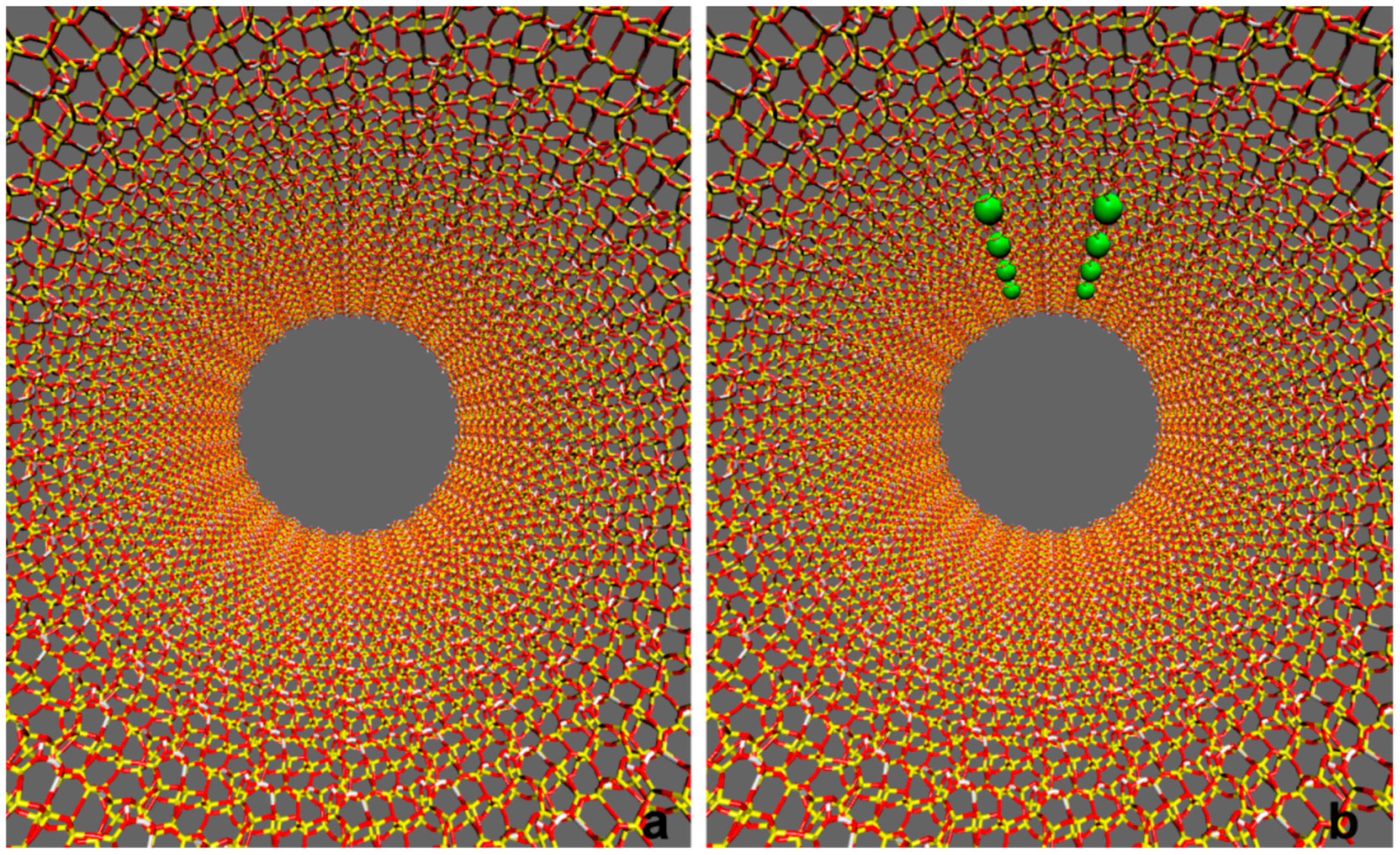
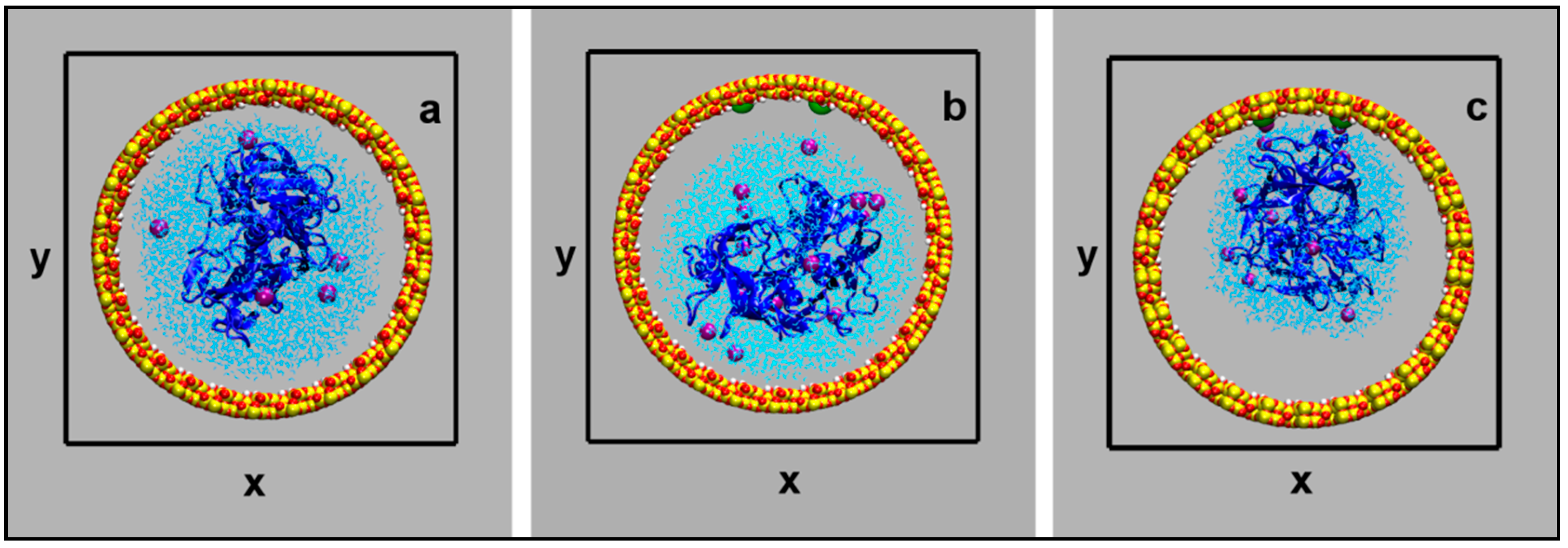
2. Results and Discussion
3. Models and Methods
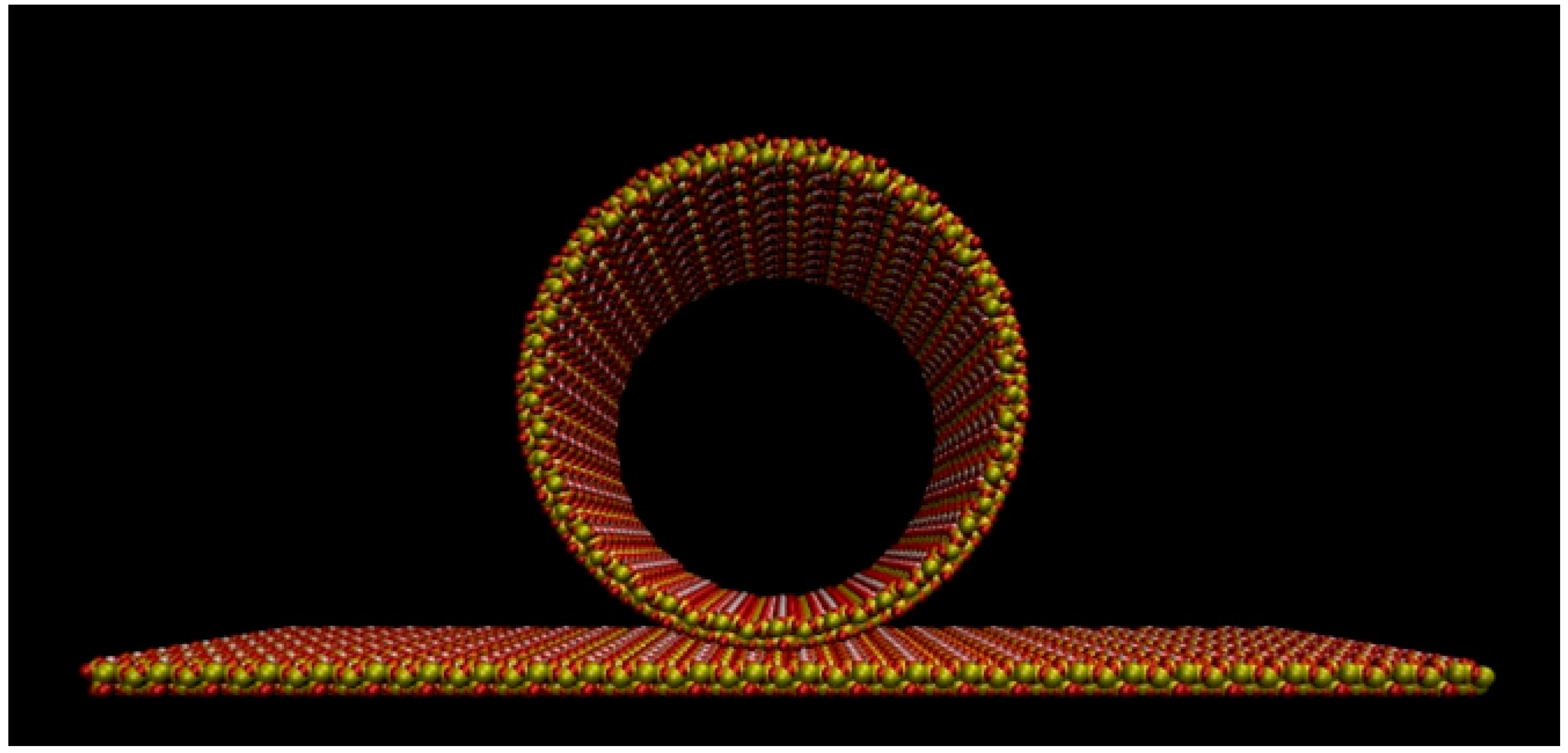
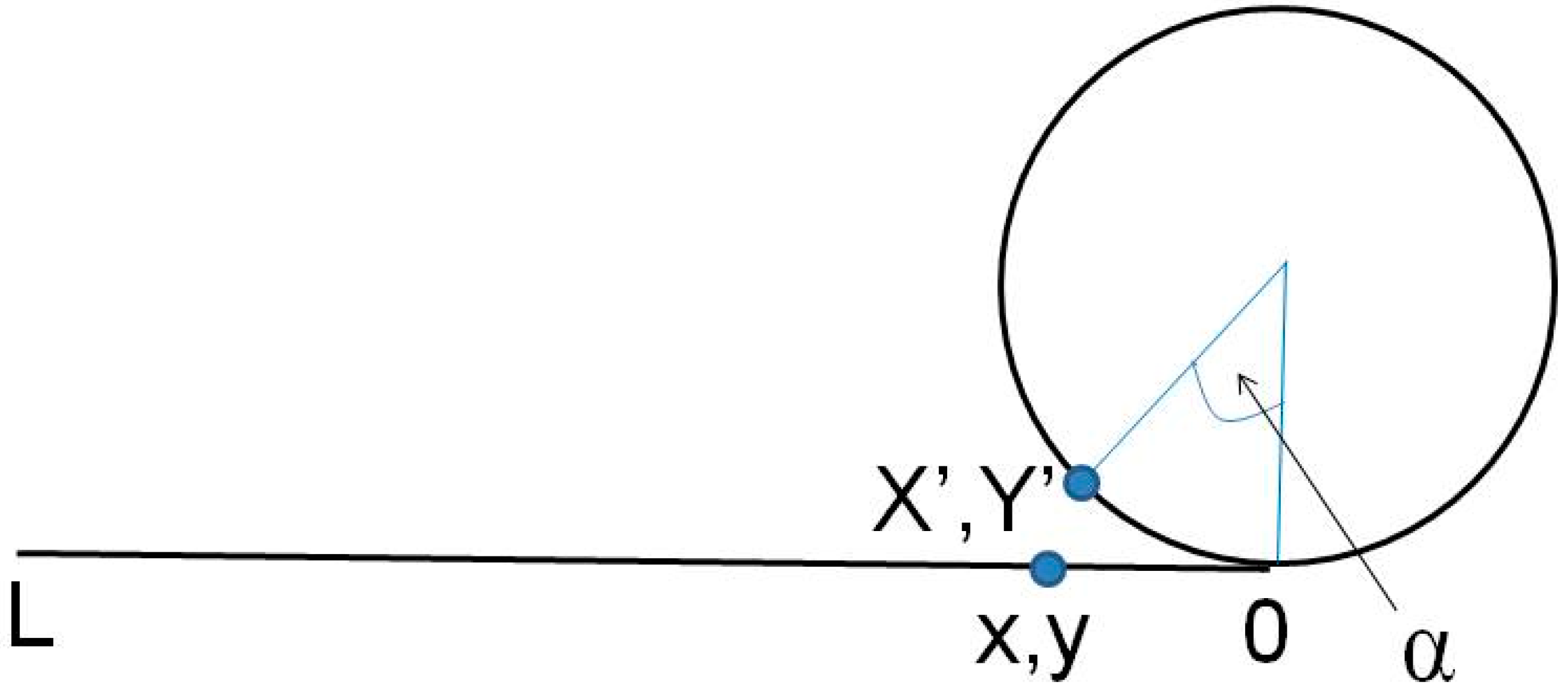
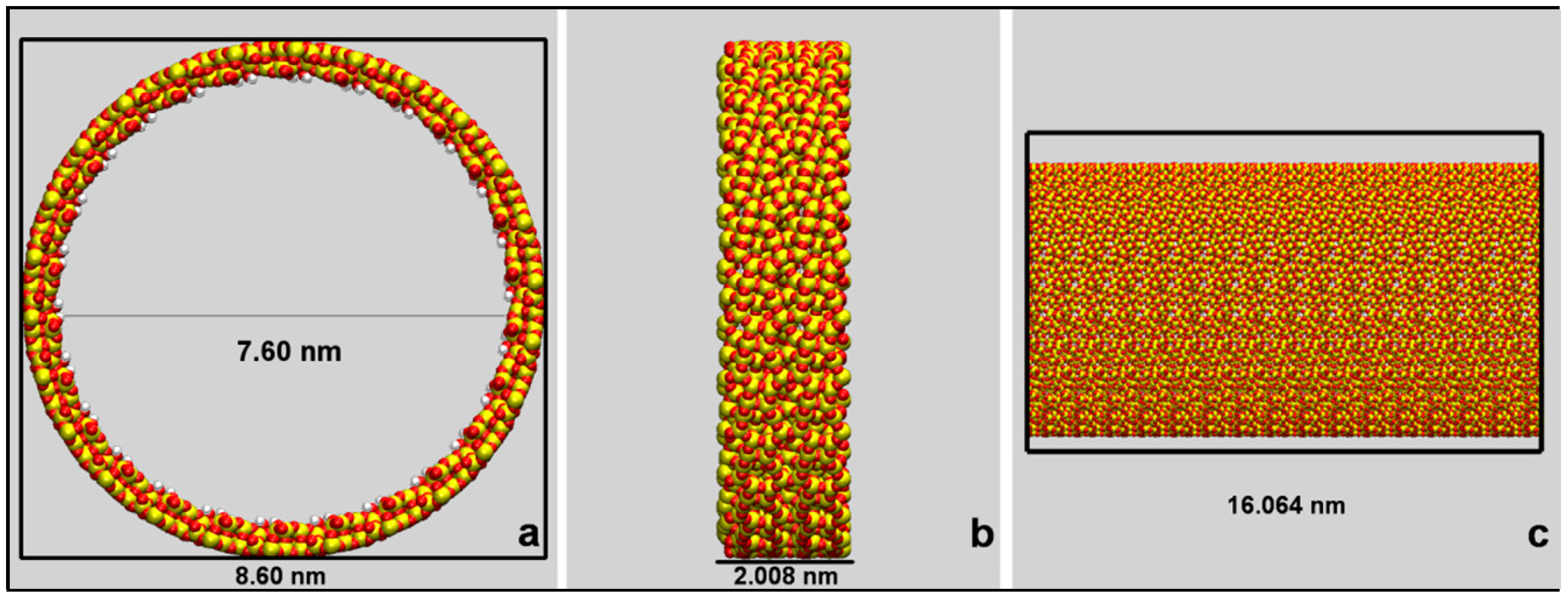
3.1. Building and Equilibration of the Mesopore
3.2. Model Building and Simulation of the Nanobiodroplet Inside the Mesopore
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Lambert, J.-F. Adsorption and Polymerization of Amino Acids on Mineral Surfaces: A Review. Orig. Life Evol. Biosph. 2008, 38, 211–242. [Google Scholar] [CrossRef] [PubMed]
- Rimola, A.; Costa, D.; Sodupe, M.; Lambert, J.-F.; Ugliengo, P. Silica Surface Features and Their Role in the Adsorption of Biomolecules: Computational Modeling and Experiments. Chem. Rev. 2013, 113, 4216–4313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabbiani, M.; Pazzi, M.; Vincenti, M.; Tabacchi, G.; Fois, E.; Martra, G. Does the Abiotic Formation of Oligopeptides on TiO 2 Nanoparticles Require Special Catalytic Sites? Apparently Not. J. Nanosci. Nanotechnol. 2018, 18, 5854–5857. [Google Scholar] [CrossRef] [PubMed]
- Tavanti, F.; Pedone, A.; Menziani, M.C. Competitive Binding of Proteins to Gold Nanoparticles Disclosed by Molecular Dynamics Simulations. J. Phys. Chem. C 2015, 119, 22172–22180. [Google Scholar] [CrossRef]
- Deyev, S.; Proshkina, G.; Ryabova, A.; Tavanti, F.; Menziani, M.C.; Eidelshtein, G.; Avishai, G.; Kotlyar, A. Synthesis, Characterization, and Selective Delivery of DARPin–Gold Nanoparticle Conjugates to Cancer Cells. Bioconjug. Chem. 2017, 28, 2569–2574. [Google Scholar] [CrossRef]
- Shaikh, S.; Nazam, N.; Rizvi, S.M.D.; Ahmad, K.; Baig, M.H.; Lee, E.J.; Choi, I.; Shaikh, S.; Nazam, N.; Rizvi, S.M.D.; et al. Mechanistic Insights into the Antimicrobial Actions of Metallic Nanoparticles and Their Implications for Multidrug Resistance. Int. J. Mol. Sci. 2019, 20, 2468. [Google Scholar] [CrossRef] [PubMed]
- Castillo, R.R.; Vallet-Regí, M. Functional Mesoporous Silica Nanocomposites: Biomedical applications and Biosafety. Int. J. Mol. Sci. 2019, 20, 929. [Google Scholar] [CrossRef]
- Cormack, A.N.; Tilocca, A. Structure and biological activity of glasses and ceramics. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2012, 370, 1271–1280. [Google Scholar] [CrossRef]
- Yu, M.; Gu, Z.; Ottewell, T.; Yu, C. Silica-based nanoparticles for therapeutic protein delivery. J. Mater. Chem. B 2017, 5, 3241–3252. [Google Scholar] [CrossRef]
- Li, Z.; Barnes, J.C.; Bosoy, A.; Stoddart, J.F.; Zink, J.I. Mesoporous silica nanoparticles in biomedical applications. Chem. Soc. Rev. 2012, 41, 2590. [Google Scholar] [CrossRef]
- Fried, D.I.; Brieler, F.J.; Fröba, M. Designing Inorganic Porous Materials for Enzyme Adsorption and Applications in Biocatalysis. ChemCatChem 2013, 5, 862–884. [Google Scholar] [CrossRef]
- Meissner, J.; Prause, A.; Di Tommaso, C.; Bharti, B.; Findenegg, G.H. Protein Immobilization in Surface-Functionalized SBA-15: Predicting the Uptake Capacity from the Pore Structure. J. Phys. Chem. C 2015, 119, 2438–2446. [Google Scholar] [CrossRef]
- Leong, J.; Seo, Y.; Chu, S.H.; Park, C.; Jeon, E.J.; Cho, S.W.; Yang, Y.Y.; Dipietro, L.A.; Kim, D.H.; Kong, H. Pore Diameter of Mesoporous Silica Modulates Oxidation of H 2 O 2 -Sensing Chromophore in a Porous Matrix. Langmuir 2018, 34, 11242–11252. [Google Scholar] [CrossRef] [PubMed]
- Carlsson, N.; Gustafsson, H.; Thörn, C.; Olsson, L.; Holmberg, K.; Åkerman, B. Enzymes immobilized in mesoporous silica: A physical-chemical perspective. Adv. Colloid Interface Sci. 2014, 205, 339–360. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Zhou, G.; Zhang, H. Synthesis, functionalization, and applications of morphology-controllable silica-based nanostructures: A review. Prog. Solid State Chem. 2016, 44, 1–19. [Google Scholar] [CrossRef]
- Mathé, C.; Devineau, S.; Aude, J.C.; Lagniel, G.; Chédin, S.; Legros, V.; Mathon, M.H.; Renault, J.P.; Pin, S.; Boulard, Y.; et al. Structural determinants for protein adsorption/non-adsorption to silica surface. PLoS ONE 2013, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Fois, E.; Gamba, A.; Tabacchi, G. Influence of silanols condensation on surface properties of micelle-templated silicas: A modelling study. Microporous Mesoporous Mater. 2008, 116, 718–722. [Google Scholar] [CrossRef]
- Solano-Umaña, V.; Vega-Baudrit, J.R. Micro, Meso and Macro Porous Materials on Medicine. J. Biomater. Nanobiotechnol. 2015, 06, 247–256. [Google Scholar] [CrossRef]
- Tabacchi, G. Supramolecular Organization in Confined Nanospaces. ChemPhysChem 2018, 19, 1249–1297. [Google Scholar] [CrossRef]
- Gigli, L.; Arletti, R.; Tabacchi, G.; Fabbiani, M.; Vitillo, J.G.; Martra, G.; Devaux, A.; Miletto, I.; Quartieri, S.; Calzaferri, G.; et al. Structure and Host–Guest Interactions of Perylene–Diimide Dyes in Zeolite L Nanochannels. J. Phys. Chem. C 2018, 122, 3401–3418. [Google Scholar] [CrossRef]
- Devaux, A.; Calzaferri, G.; Belser, P.; Cao, P.; Brühwiler, D.; Kunzmann, A. Efficient and robust host-guest antenna composite for light harvesting. Chem. Mater. 2014, 26, 6878–6885. [Google Scholar] [CrossRef]
- Zucchetto, N.; Brühwiler, D. Strategies for Localizing Multiple Functional Groups in Mesoporous Silica Particles through a One-Pot Synthesis. Chem. Mater. 2018, 30, 7280–7286. [Google Scholar] [CrossRef] [Green Version]
- Tabacchi, G.; Fois, E.; Calzaferri, G. Structure of Nanochannel Entrances in Stopcock-Functionalized Zeolite L Composites. Angew. Chem. Int. Ed. 2015, 54, 11112–11116. [Google Scholar] [CrossRef] [PubMed]
- Bagnall, A.J.; Santana Vega, M.; Martinelli, J.; Djanashvili, K.; Cucinotta, F. Mesoscopic FRET Antenna Materials by Self-Assembling Iridium(III) Complexes and BODIPY Dyes. Chem. Eur. J. 2018, 24, 11992–11999. [Google Scholar] [CrossRef] [PubMed]
- Kehr, N.S. Microcontact Printing of (Bio)Molecules on Self-Assembled Monolayers of Zeolites L and Surface Mediated Drug Delivery. Adv. Porous Mater. 2018, 6, 19–27. [Google Scholar] [CrossRef]
- Bertucci, A.; Lülf, H.; Septiadi, D.; Manicardi, A.; Corradini, R.; De Cola, L. Intracellular Delivery of Peptide Nucleic Acid and Organic Molecules Using Zeolite-L Nanocrystals. Adv. Healthc. Mater. 2014, 3, 1812–1817. [Google Scholar] [CrossRef] [PubMed]
- Devaux, A.; Cucinotta, F.; Kehr, S.; Cola, L. De Functionalization and Assembling of Inorganic Nanocontainers for Optical and Biomedical Applications. In Functional Supramolecular Architectures; WILEY-VCH Verlag & Co. KGaA: Weinheim, Germany, 2014; pp. 281–342. ISBN 9783527689897. [Google Scholar]
- Calzaferri, G. Nanochannels: Hosts for the supramolecular organization of molecules and complexes. Langmuir 2012, 28, 6216–6231. [Google Scholar] [CrossRef]
- Gößl, D.; Singer, H.; Chiu, H.-Y.; Schmidt, A.; Lichtnecker, M.; Engelke, H.; Bein, T. Highly active enzymes immobilized in large pore colloidal mesoporous silica nanoparticles. New J. Chem. 2019, 43, 1671–1680. [Google Scholar] [CrossRef]
- Onida, B.; Bonelli, B.; Flora, L.; Geobaldo, F.; Arean, C.O.; Garrone, E. Permeability of micelles in surfactant-containing MCM-41 silica as monitored by embedded dye molecules. Chem. Commun. (Camb). 2001, 0, 2216–2217. [Google Scholar] [CrossRef]
- Manyar, H.G.; Gianotti, E.; Sakamoto, Y.; Terasaki, O.; Coluccia, S.; Tumbiolo, S. Active Biocatalysts Based on Pepsin Immobilized in Mesoporous SBA-15. J. Phys. Chem. C 2008, 112, 18110–18116. [Google Scholar] [CrossRef]
- Thörn, C.; Udatha, D.B.R.K.G.; Zhou, H.; Christakopoulos, P.; Topakas, E.; Olsson, L. Understanding the pH-dependent immobilization efficacy of feruloyl esterase-C on mesoporous silica and its structure–activity changes. J. Mol. Catal. B Enzym. 2013, 93, 65–72. [Google Scholar] [CrossRef]
- Meissner, J.; Prause, A.; Bharti, B.; Findenegg, G.H. Characterization of protein adsorption onto silica nanoparticles: Influence of pH and ionic strength. Colloid Polym. Sci. 2015, 293, 3381–3391. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhou, G.; Wu, C.; Li, T.; Song, H. Improving adsorption and activation of the lipase immobilized in amino-functionalized ordered mesoporous SBA-15. Solid State Sci. 2011, 13, 867–874. [Google Scholar] [CrossRef]
- Moerz, S.T.; Huber, P. PH-Dependent Selective Protein Adsorption into Mesoporous Silica. J. Phys. Chem. C 2015, 119, 27072–27079. [Google Scholar] [CrossRef]
- Lundqvist, M.; Sethson, I.; Jonsson, B.H. Protein adsorption onto silica nanoparticles: Conformational changes depend on the particles’ curvature and the protein stability. Langmuir 2004, 20, 10639–10647. [Google Scholar] [CrossRef]
- Jarisz, T.A.; Lane, S.; Gozdzialski, L.; Hore, D.K. Ions, metabolites, and cells: Water as a reporter of surface conditions during bacterial growth. J. Chem. Phys. 2018, 148, 222825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mogharabi-Manzari, M.; Amini, M.; Abdollahi, M.; Khoobi, M.; Bagherzadeh, G.; Faramarzi, M.A. Co-immobilization of Laccase and TEMPO in the Compartments of Mesoporous Silica for a Green and One-Pot Cascade Synthesis of Coumarins by Knoevenagel Condensation. ChemCatChem 2018, 10, 1542–1546. [Google Scholar] [CrossRef]
- El-Baba, T.J.; Fuller, D.R.; Woodall, D.W.; Raab, S.A.; Conant, C.R.; Dilger, J.M.; Toker, Y.; Williams, E.R.; Russell, D.H.; Clemmer, D.E. Melting proteins confined in nanodroplets with 10.6 μm light provides clues about early steps of denaturation. Chem. Commun. 2018, 54, 3270–3273. [Google Scholar] [CrossRef]
- Cohavi, O.; Corni, S.; de Rienzo, F.; di Felice, R.; Gottschalk, K.E.; Hoefling, M.; Kokh, D.; Molinari, E.; Schreiber, G.; Vaskevich, A.; et al. Protein-surface interactions: Challenging experiments and computations. J. Mol. Recognit. 2010, 23, 259–262. [Google Scholar] [CrossRef]
- Trachta, M.; Bludský, O.; Rubeš, M. The interaction of proteins with silica surfaces. Part II: Free energies of capped amino acids. Comput. Theor. Chem. 2019, 1148, 38–43. [Google Scholar] [CrossRef]
- Corno, M.; Delle Piane, M.; Choquet, P.; Ugliengo, P. Models for biomedical interfaces: A computational study of quinone-functionalized amorphous silica surface features. Phys. Chem. Chem. Phys. 2017, 19, 7793–7806. [Google Scholar] [CrossRef]
- Oliver, D.; Michaelis, M.; Heinz, H.; Volkov, V.V.; Perry, C.C. From phage display to structure: An interplay of enthalpy and entropy in the binding of the LDHSLHS polypeptide to silica. Phys. Chem. Chem. Phys. 2019, 21, 4663–4672. [Google Scholar] [CrossRef]
- Jayanthi, S.; Kababya, S.; Schmidt, A.; Vega, S. Deuterium MAS NMR and Local Molecular Dynamic Model to Study Adsorption–Desorption Kinetics of a Dipeptide at the Inner Surfaces of SBA-15. J. Phys. Chem. C 2016, 120, 2797–2806. [Google Scholar] [CrossRef]
- Kubiak, K.; Mulheran, P.A. Molecular Dynamics Simulations of Hen Egg White Lysozyme Adsorption at a Charged Solid Surface. J. Phys. Chem. B 2009, 113, 12189–12200. [Google Scholar] [CrossRef]
- Sun, X.; Feng, Z.; Zhang, L.; Hou, T.; Li, Y. The Selective Interaction between Silica Nanoparticles and Enzymes from Molecular Dynamics Simulations. PLoS ONE 2014, 9, e107696. [Google Scholar] [CrossRef]
- He, J.; Wu, M.; Feng, X.; Shao, X.; Cai, W. Immobilization of papain on nanoporous silica. RSC Adv. 2014, 4, 13304–13312. [Google Scholar] [CrossRef]
- Raffaini, G.; Ganazzoli, F.; Stuart, S.J.; Latour, R.A.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. Protein adsorption on the hydrophilic surface of a glassy polymer: A computer simulation study. Phys. Chem. Chem. Phys. 2006, 8, 2765. [Google Scholar] [CrossRef]
- Artali, R.; Pra, A.D.; Foresti, E.; Lesci, I.G.; Roveri, N.; Sabatino, P. Adsorption of human serum albumin on the chrysotile surface: A molecular dynamics and spectroscopic investigation. J. R. Soc. Interface 2008, 5, 273–283. [Google Scholar] [CrossRef]
- Liamas, E.; Kubiak-Ossowska, K.; Black, R.; Thomas, O.; Zhang, Z.; Mulheran, P.; Liamas, E.; Kubiak-Ossowska, K.; Black, R.A.; Thomas, O.R.T.; et al. Adsorption of Fibronectin Fragment on Surfaces Using Fully Atomistic Molecular Dynamics Simulations. Int. J. Mol. Sci. 2018, 19, 3321. [Google Scholar] [CrossRef]
- Coasne, B.; Ugliengo, P. Atomistic model of micelle-templated mesoporous silicas: Structural, morphological, and adsorption properties. Langmuir 2012, 28, 11131–11141. [Google Scholar] [CrossRef]
- Delle Piane, M.; Corno, M.; Pedone, A.; Dovesi, R.; Ugliengo, P. Large-scale B3LYP simulations of ibuprofen adsorbed in MCM-41 mesoporous silica as drug delivery system. J. Phys. Chem. C 2014, 118, 26737–26749. [Google Scholar] [CrossRef]
- Pedone, A.; Bloino, J.; Barone, V. Role of Host–Guest Interactions in Tuning the Optical Properties of Coumarin Derivatives Incorporated in MCM-41: A TD-DFT Investigation. J. Phys. Chem. C 2012, 116, 17807–17818. [Google Scholar] [CrossRef]
- Trachta, M.; Bludský, O.; Rubeš, M. The interaction of proteins with silica surfaces. Part I: Ab initio modeling. Comput. Theor. Chem. 2017, 1117, 100–107. [Google Scholar] [CrossRef]
- Folliet, N.; Gervais, C.; Costa, D.; Laurent, G.; Babonneau, F.; Stievano, L.; Lambert, J.-F.; Tielens, F. A Molecular Picture of the Adsorption of Glycine in Mesoporous Silica through NMR Experiments Combined with DFT-D Calculations. J. Phys. Chem. C 2013, 117, 4104–4114. [Google Scholar] [CrossRef]
- Rimola, A.; Civalleri, B.; Ugliengo, P. Neutral vs Zwitterionic Glycine Forms at the Water/Silica Interface: Structure, Energies, and Vibrational Features from B3LYP Periodic Simulations. Langmuir 2008, 24, 14027–14034. [Google Scholar] [CrossRef]
- Fois, E.; Tabacchi, G.; Calzaferri, G. Orientation and order of xanthene dyes in the one-dimensional channels of zeolite L: Bridging the gap between experimental data and molecular behavior. J. Phys. Chem. C 2012, 116, 16784–16799. [Google Scholar] [CrossRef]
- Zhou, X.; Wesolowski, T.A.; Tabacchi, G.; Fois, E.; Calzaferri, G.; Devaux, A. First-principles simulation of the absorption bands of fluorenone in zeolite L. Phys. Chem. Chem. Phys. 2013, 15, 159–167. [Google Scholar] [CrossRef]
- Tabacchi, G.; Calzaferri, G.; Fois, E. One-dimensional self-assembly of perylene-diimide dyes by unidirectional transit of zeolite channel openings. Chem. Commun. 2016, 52, 11195–11198. [Google Scholar] [CrossRef] [Green Version]
- Tilocca, A.; Fois, E. The color and stability of maya blue: TDDFT calculations. J. Phys. Chem. C 2009, 113, 8683–8687. [Google Scholar] [CrossRef]
- Kim, J.; Grate, J.W.; Wang, P. Nanostructures for enzyme stabilization. Chem. Eng. Sci. 2006, 61, 1017–1026. [Google Scholar] [CrossRef]
- Sielecki, A.R.; Fedorov, A.A.; Boodhoo, A.; Andreeva, N.S.; James, M.N.G. Molecular and crystal structures of monoclinic porcine pepsin refined at 1.8A° resolution. J. Mol. Biol. 1990, 214, 143–170. [Google Scholar] [CrossRef]
- Giussani, L.; Fois, E.; Gianotti, E.; Tabacchi, G.; Gamba, A.; Coluccia, S. The determination of pepsin dimensions at different pH values: A simulation study. Nuovo Cim. della Soc. Ital. di Fis. B 2008, 123, 1477–1483. [Google Scholar] [CrossRef]
- Giussani, L.; Fois, E.; Gianotti, E.; Tabacchi, G.; Gamba, A.; Coluccia, S. On the compatibility criteria for protein encapsulation inside mesoporous materials. ChemPhysChem 2010, 11, 1757–1762. [Google Scholar] [CrossRef]
- Malek, S.M.A.; Poole, P.H.; Saika-Voivod, I. Thermodynamic and structural anomalies of water nanodroplets. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef]
- Pillai, R.; Borg, M.K.; Reese, J.M. Dynamics of Nanodroplets on Vibrating Surfaces. Langmuir 2018, 34, 11898–11904. [Google Scholar] [CrossRef] [Green Version]
- Fois, E.; Tabacchi, G. Water in zeolite L and its MOF mimic. Zeitschrift fur Krist. Cryst. Mater. 2019. [Google Scholar] [CrossRef]
- Fraux, G.; Coudert, F.X.; Boutin, A.; Fuchs, A.H. Forced intrusion of water and aqueous solutions in microporous materials: From fundamental thermodynamics to energy storage devices. Chem. Soc. Rev. 2017, 46, 7421–7437. [Google Scholar] [CrossRef]
- Arletti, R.; Fois, E.; Gigli, L.; Vezzalini, G.; Quartieri, S.; Tabacchi, G. Irreversible Conversion of a Water–Ethanol Solution into an Organized Two-Dimensional Network of Alternating Supramolecular Units in a Hydrophobic Zeolite under Pressure. Angew. Chem. Int. Ed. 2017, 56, 2105–2109. [Google Scholar] [CrossRef]
- Fischer, M. First-Principles Study of AlPO4-H3, a Hydrated Aluminophosphate Zeotype Containing Two Different Types of Adsorbed Water Molecules. Molecules 2019, 24, 922. [Google Scholar] [CrossRef]
- Gatta, G.D.; Lotti, P.; Tabacchi, G. The effect of pressure on open-framework silicates: Elastic behaviour and crystal–fluid interaction. Phys. Chem. Miner. 2018, 45, 115–138. [Google Scholar] [CrossRef]
- Muñoz-Santiburcio, D.; Marx, D. Nanoconfinement in Slit Pores Enhances Water Self-Dissociation. Phys. Rev. Lett. 2017, 119, 056002. [Google Scholar] [CrossRef] [PubMed]
- Scalfi, L.; Fraux, G.; Boutin, A.; Coudert, F.X. Structure and Dynamics of Water Confined in Imogolite Nanotubes. Langmuir 2018, 34, 6748–6756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coudert, F.-X.; Cailliez, F.; Vuilleumier, R.; Fuchs, A.H.; Boutin, A. Water nanodroplets confined in zeolite pores. Faraday Discuss. 2009, 141, 377–398. [Google Scholar] [CrossRef]
- Gallo, P.; Rovere, M.; Chen, S.-H. Anomalous dynamics of water confined in MCM-41 at different hydrations. J. Phys. Condens. Matter 2010, 22, 284102. [Google Scholar] [CrossRef] [PubMed]
- Coudert, F.-X.; Vuilleumier, R.; Boutin, A. Dipole Moment, Hydrogen Bonding and IR Spectrum of Confined Water. ChemPhysChem 2006, 7, 2464–2467. [Google Scholar] [CrossRef] [Green Version]
- Demontis, P.; Gulìn-Gonzàlez, J.; Jobic, H.; Masia, M.; Sale, R.; Suffritti, G.B. Dynamical properties of confined water nanoclusters: Simulation study of hydrated zeolite NaA: Structural and vibrational properties. ACS Nano 2008, 2, 1603–1614. [Google Scholar] [CrossRef]
- Suffritti, G.B.; Demontis, P.; Gulín-Gonzlez, J.; Masia, M. Computer simulations of dynamic crossover phenomena in nanoconfined water. J. Phys. Condens. Matter 2012, 24. [Google Scholar] [CrossRef]
- Wang, C.H.; Bai, P.; Siepmann, J.I.; Clark, A.E. Deconstructing hydrogen-bond networks in confined nanoporous materials: Implications for alcohol-water separation. J. Phys. Chem. C 2014, 118, 19723–19732. [Google Scholar] [CrossRef]
- Fois, E.; Gamba, A.; Medici, C.; Tabacchi, G.; Quartieri, S.; Mazzucato, E.; Arletti, R.; Vezzalini, G.; Dmitriev, V. High pressure deformation mechanism of Li-ABW: Synchrotron XRPD study and ab initio molecular dynamics simulations. Microporous Mesoporous Mater. 2008, 115, 267–280. [Google Scholar] [CrossRef]
- Cailliez, F.; Trzpit, M.; Soulard, M.; Demachy, I.; Boutin, A.; Patarin, J.; Fuchs, A.H. Thermodynamics of water intrusion in nanoporous hydrophobic solids. Phys. Chem. Chem. Phys. 2008, 10, 4817. [Google Scholar] [CrossRef]
- Demontis, P.; Gulín-González, J.; Masia, M.; Suffritti, G.B. The behaviour of water confined in zeolites: Molecular dynamics simulations versus experiment. J. Phys. Condens. Matter 2010, 22, 284106. [Google Scholar] [CrossRef] [PubMed]
- Conant, C.R.; Fuller, D.R.; Zhang, Z.; Woodall, D.W.; Russell, D.H.; Clemmer, D.E. Substance P in the Gas Phase: Conformational Changes and Dissociations Induced by Collisional Activation in a Drift Tube. J. Am. Soc. Mass Spectrom. 2019, 30, 932–945. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulou, E.; Megaridis, C.M.; Walther, J.H.; Koumoutsakos, P. Ultrafast Propulsion of Water Nanodroplets on Patterned Graphene. ACS Nano 2019, acsnano.9b00252. [Google Scholar] [CrossRef] [PubMed]
- Giussani, L.; Tabacchi, G.; Gianotti, E.; Coluccia, S.; Fois, E. Disentangling protein-silica interactions. Philos. Trans. A. Math. Phys. Eng. Sci. 2012, 370, 1463–1477. [Google Scholar] [CrossRef] [PubMed]
- Okoniewska, M.; Tanaka, T.; Yada, R.Y. The pepsin residue glycine-76 contributes to active-site loop flexibility and participates in catalysis. Biochem. J. 2000, 349, 169–177. [Google Scholar] [CrossRef]
- Deere, J.; Magner, E.; Wall, J.G.; Hodnett, B.K. Mechanistic and structural features of protein adsorption onto mesoporous silicates. J. Phys. Chem. B 2002, 106, 7340–7347. [Google Scholar] [CrossRef]
- Izadi, S.; Onufriev, A.V. Accuracy limit of rigid 3-point water models. J. Chem. Phys. 2016, 145, 074501. [Google Scholar] [CrossRef] [Green Version]
- Shabane, P.S.; Izadi, S.; Onufriev, A.V. General Purpose Water Model Can Improve Atomistic Simulations of Intrinsically Disordered Proteins. J. Chem. Theory Comput. 2019, 15, 2620–2634. [Google Scholar] [CrossRef]
- Javanainen, M.; Lamberg, A.; Cwiklik, L.; Vattulainen, I.; Ollila, O.H.S. Atomistic Model for Nearly Quantitative Simulations of Langmuir Monolayers. Langmuir 2018, 34, 2565–2572. [Google Scholar] [CrossRef]
- Fois, E.; Gamba, A.; Tabacchi, G.; Coluccia, S.; Martra, G. Ab Initio Study of Defect Sites at the Inner Surfaces of Mesoporous Silicas. J. Phys. Chem. B 2003, 107, 10767–10772. [Google Scholar] [CrossRef]
- Tabacchi, G.; Gianotti, E.; Fois, E.; Martra, G.; Marchese, L.; Coluccia, S.; Gamba, A. Understanding the vibrational and electronic features of Ti(IV) sites in mesoporous silicas by integrated Ab initio and spectroscopic investigations. J. Phys. Chem. C 2007, 111, 4946–4955. [Google Scholar] [CrossRef]
- Fois, E.; Gamba, A.; Tabacchi, G.; Coluccia, S.; Martra, G. Properties of defect centres on nanothick silica layers: An ab initio investigation. J. Porous Mater. 2007, 14, 339–347. [Google Scholar] [CrossRef]
- Fois, E.; Tabacchi, G.; Barreca, D.; Gasparotto, A.; Tondello, E. “Hot” surface activation of molecular complexes: Insight from modeling studies. Angew. Chem. Int. Ed. 2010, 49, 1944–1948. [Google Scholar] [CrossRef] [PubMed]
- Tabacchi, G.; Fois, E.; Barreca, D.; Carraro, G.; Gasparotto, A.; Maccato, C. Modeling The First Activation Stages of the Fe(hfa)2TMEDA CVD Precursor on a Heated Growth Surface. In Advanced Processing and Manufacturing Technologies for Nanostructured and Multifunctional Materials II; Ohji, T., Singh, M., Halbig, M., Eds.; Ceramic Engineering and Science Proceedings; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2016; pp. 83–90. ISBN 9781119211662. [Google Scholar] [CrossRef]
- Demontis, P.; Suffritti, G.B.; Quartieri, S.; Gamba, A.; Fois, E.S. Molecular dynamics studies on zeolites. Part 5.—Discussion of the structural changes of silicalite. J. Chem. Soc. Faraday Trans. 1991, 87, 1657–1663. [Google Scholar] [CrossRef]
- Allolio, C.; Klameth, F.; Vogel, M.; Sebastiani, D. Ab initio H2O in realistic hydrophilic confinement. ChemPhysChem 2014, 15, 3955–3962. [Google Scholar] [CrossRef] [PubMed]
- Ottaviani, M.F.; Galarneau, A.; Desplantier-Giscard, D.; Di Renzo, F.; Fajula, F. EPR investigations on the formation of micelle-templated silica. Microporous Mesoporous Mater. 2001, 44–45, 1–8. [Google Scholar] [CrossRef]
- MacKerell, A.D.; Bashford, D.; Bellott, M.; Dunbrack, R.L.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins †. J. Phys. Chem. B 1998, 102, 3586–3616. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Batcho, P.F.; Case, D.A.; Schlick, T. Optimized particle-mesh Ewald/multiple-time step integration for molecular dynamics simulations. J. Chem. Phys. 2001, 115, 4003–4018. [Google Scholar] [CrossRef] [Green Version]
- Onufriev, A.V.; Izadi, S. Water models for biomolecular simulations. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2018, 8, e1347. [Google Scholar] [CrossRef]
- Cicu, P.; Demontis, P.; Spanu, S.; Suffritti, G.B.; Tilocca, A. Electric-field-dependent empirical potentials for molecules and crystals: A first application to flexible water molecule adsorbed in zeolites. J. Chem. Phys. 2000, 112, 8267–8278. [Google Scholar] [CrossRef] [Green Version]
- Maerzke, K.A.; McGrath, M.J.; William Kuo, I.F.; Tabacchi, G.; Siepmann, J.I.; Mundy, C.J. Vapor-liquid phase equilibria of water modelled by a Kim-Gordon potential. Chem. Phys. Lett. 2009, 479, 60–64. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]


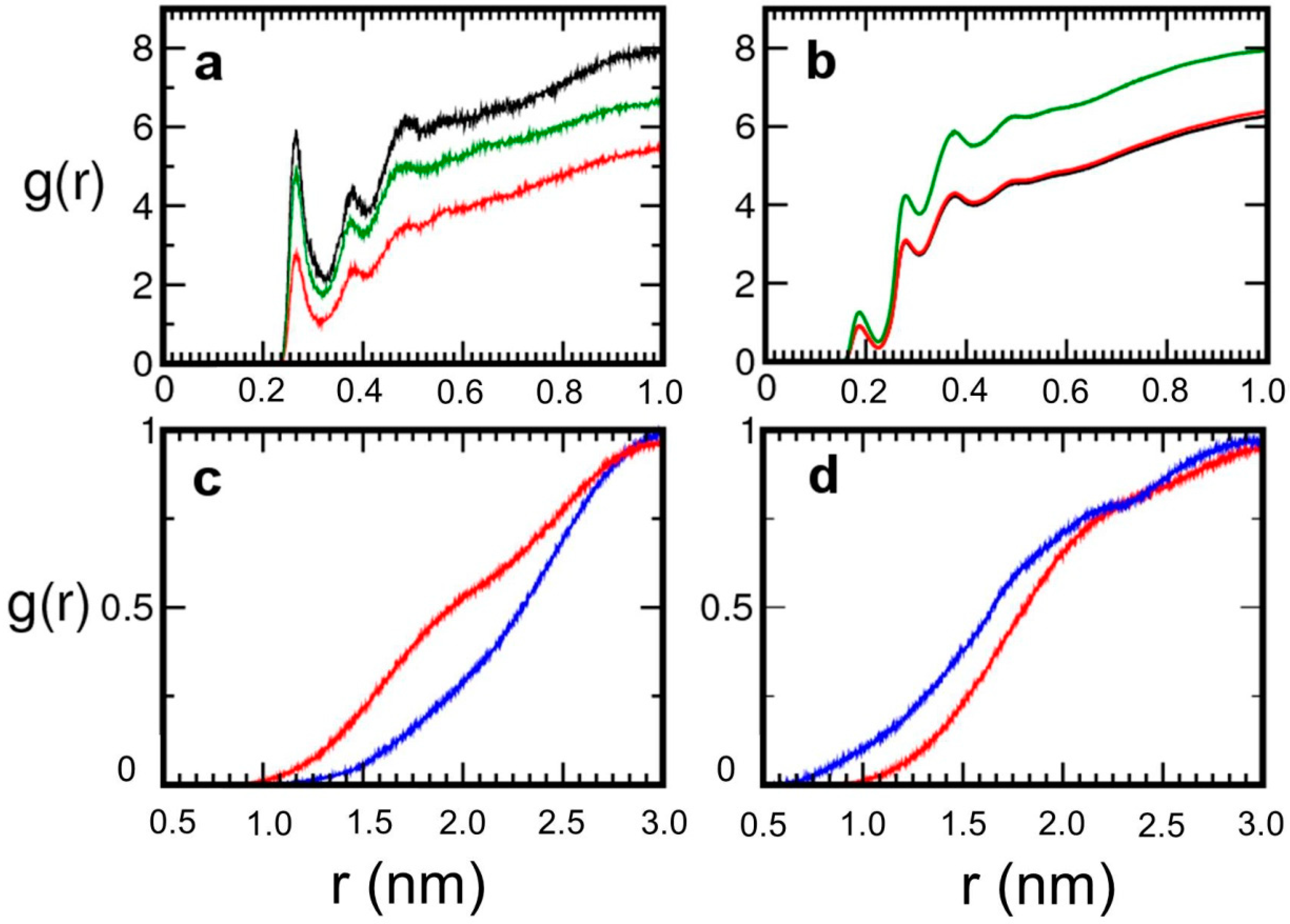
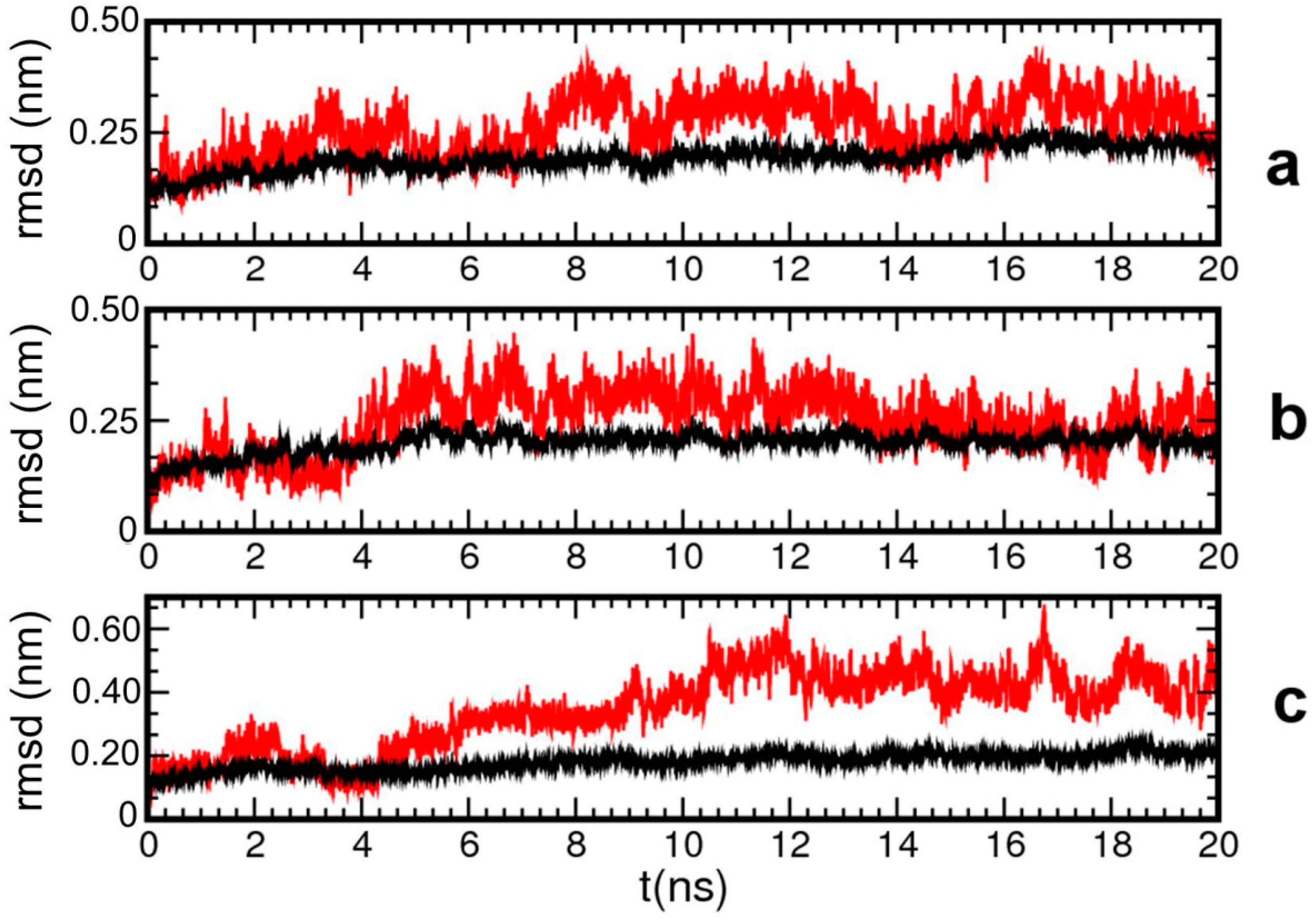
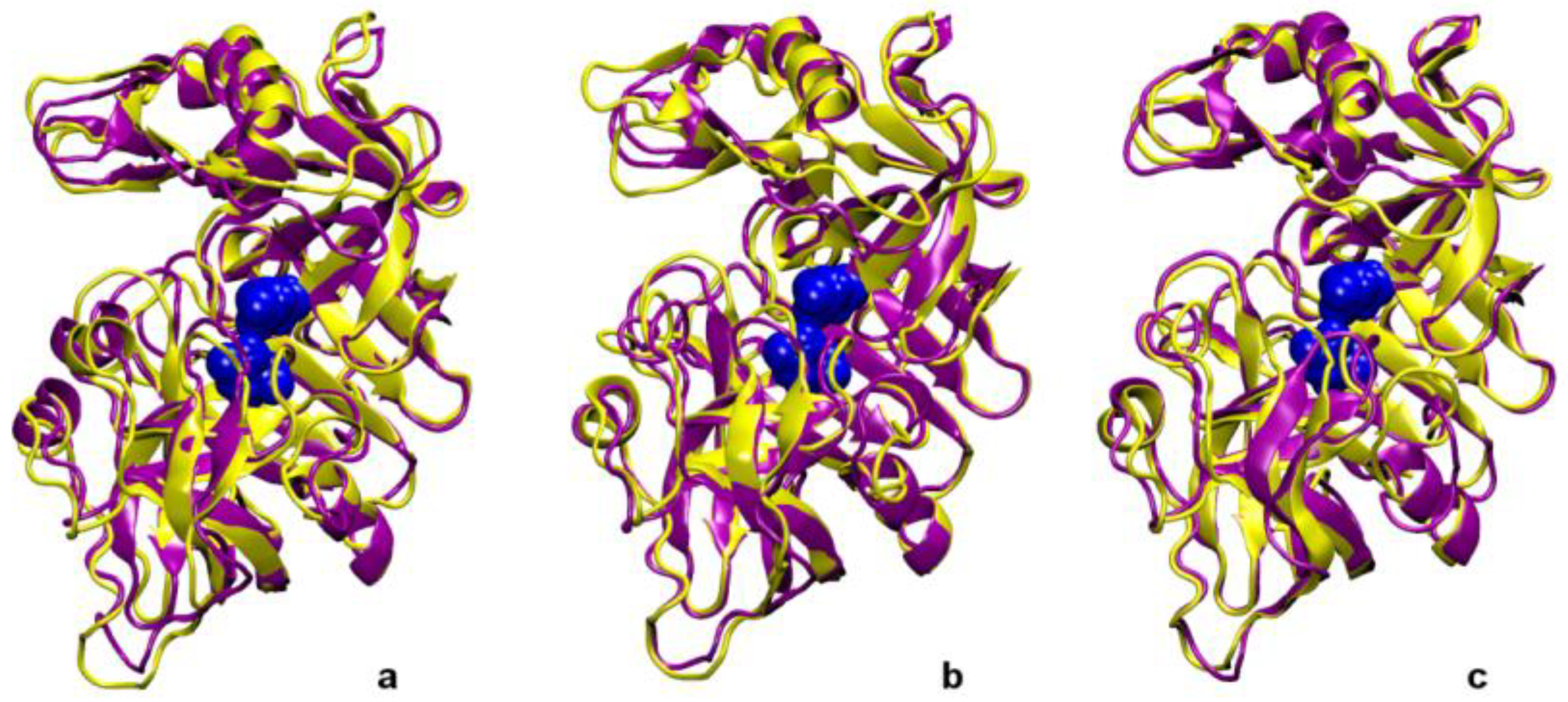


| Pair | Model 1 | Model 2 | Model 3 |
|---|---|---|---|
| Pep–water | −24,230 (565) | −23,882 (611) | −23,326 (624) |
| Pep–silica channel | −54 (4) | 268 (92) | 469 (59) |
| Pep–K+ | −1665 (351) | −2548 (377) | −3234 (389) |
| Pep–K+ per ion | −276 (59) | −184 (25) | −230 (29) |
| Water–silica channel | −80 (21) | −255 (293) | −151 (226) |
| Water–K+ | −2946 (343) | −6418 (552) | −5724 (477) |
| Water–K+ per ion a | −490 (59) | −460 (38) | −410 (33) |
| Silica channel–K+ | −121 (54) | −2536 (46) | −3247 (247) |
| Pep–(silica channel + water + K+) | −25,949 (469) | −26,376 (444) | −25,882 (510) |
| Silica channel–(pep + water + K+) | −80 (21) | −2561 (96) | −2632 (84) |
| System | Asp215-Asp32 | Gly76-Asp215 | Asp215-Surface | Ser61-Glu202 |
|---|---|---|---|---|
| Model 1 | 0.486 ± 0.045 | 0.880 ± 0.155 | 3.444 ± 0.134 | 6.388 ± 0.104 |
| Model 2 | 0.520 ± 0.077 | 1.060 ± 0.206 | 2.773 ± 0127 | 6.430 ± 0.096 |
| Model 3 | 0.522 ± 0.089 | 1.187 ± 0.165 | 2.927 ± 0096 | 6.300 ± 0.140 |
| Pep–crystal structure | 0.510 | 1.040 | - | 59 |
| Atom-Atom Pair | ε (kJ/mol) | Rmin (nm) |
|---|---|---|
| Si–Si | 0 | 0 |
| Si–O | 109.04 | 0.167 |
| O–O | 1.979 | 0.328 |
| O–H | 0.946 | 0.145 |
| Si–H | 0.0874 | 0.135 |
| H–H | 0.0249 | 0.101 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giussani, L.; Tabacchi, G.; Coluccia, S.; Fois, E. Confining a Protein-Containing Water Nanodroplet inside Silica Nanochannels. Int. J. Mol. Sci. 2019, 20, 2965. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20122965
Giussani L, Tabacchi G, Coluccia S, Fois E. Confining a Protein-Containing Water Nanodroplet inside Silica Nanochannels. International Journal of Molecular Sciences. 2019; 20(12):2965. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20122965
Chicago/Turabian StyleGiussani, Lara, Gloria Tabacchi, Salvatore Coluccia, and Ettore Fois. 2019. "Confining a Protein-Containing Water Nanodroplet inside Silica Nanochannels" International Journal of Molecular Sciences 20, no. 12: 2965. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20122965