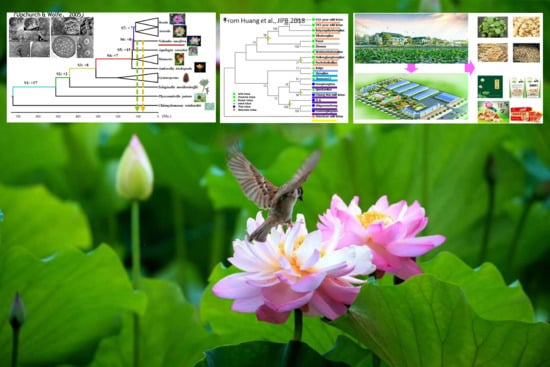
The Latest Studies on Lotus (Nelumbo nucifera)-an Emerging Horticultural Model Plant
Abstract
:1. Introduction
2. Phylogeny and Genomic Studies
3. Unique Properties of Lotus
4. Genetic and Molecular Studies on the Horticultural Traits of the Lotus
4.1. The Flower of Lotus
4.2. Rhizome and Seeds
4.3. Secondary Metabolites and Medicinal Usage of Lotus
4.4. Studies on the Establishment of Lotus Regeneration and Transformation System
5. Conclusions and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AFLP | Amplified Fragment Length Polymorphism |
| AGL | AGAMOUS-like |
| ANN | Annexin |
| ANS | anthocyanin synthase |
| AOX | alternative oxidases |
| APG | The Angiosperm Phylogeny Group |
| AS | alternative splicing |
| BIA | benzylisoquinoline alkaloid |
| CCA | CIRCADIAN CLOCK ASSOCIATED |
| CER | ECERIFERUM |
| CHS | Chalcone synthase |
| CO-LIKE | CONSTANS-like |
| COP | CONSTITUTIVELY PHOTOMORPHOGENIC |
| Cpn | Chaperonin |
| DFR | dihydroflavonol 4-reductase |
| EF | elongation factor |
| HSP | Heat shock protein |
| FT | FLOWERING LOCUS T |
| GAI | gibberellic acid insensitive |
| ISSR | inter-simple sequence repeat |
| LHY | LATE ELONGATED HYPOCOTYL |
| MADS-box | MINICHROMOSOME MAINTENANCE 1 (MCM1), AGAMOUS (AG), DEFICIENS (DEF), and SERUM RESPONSE FACTOR (SRF) domain |
| PIMT | Protein L-isoaspartyl methyltransferase |
| PRX | Peroxiredoxin |
| PUMP | plant uncoupling mitochondrial protein |
| RAPD | Random Amplified Polymorphic DNA |
| SNP | single nucleotide polymorphism |
| SOD | superoxide demutase |
| SRAP | Sequence—related amplified polymorphism |
| SSR | Simple Sequence Repeats |
| TE | transposable element |
| VIN3 | vernalization |
References
- Wang, Q.; Zhang, X. Colored Illustration of Lotus Cultivars in China; China Forestry Publishing House: Beijing, China, 2005. [Google Scholar]
- Zhang, X.; Chen, L.; Wang, Q. New Lotus Flower Cultivars in China; China Forestry Publishing House: Beijing, China, 2011. [Google Scholar]
- Zhang, Y.; Lu, X.; Zeng, S.; Huang, X.; Guo, Z.; Zheng, Y.; Tian, Y.; Zheng, B. Nutritional composition, physiological functions and processing of lotus (Nelumbo nucifera Gaertn.) seeds: A review. Phytochem. Rev. 2015, 14, 321–334. [Google Scholar] [CrossRef]
- Li, Y.; Smith, T.; Svetlana, P.; Yang, J.; Jin, J.-H.; Li, C.-S. Paleobiogeography of the lotus plant (Nelumbonaceae: Nelumbo) and its bearing on the paleoclimatic changes. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2014, 399, 284–293. [Google Scholar] [CrossRef]
- Shen-Miller, J. Sacred lotus, the long-living fruits of China Antique. Seed Sci. Res. 2002, 12, 131–143. [Google Scholar] [CrossRef] [Green Version]
- Ming, R.; Vanburen, R.; Liu, Y.; Yang, M.; Han, Y.; Li, L.T.; Zhang, Q.; Kim, M.J.; Schatz, M.C.; Campbell, M. Genome of the long-living sacred lotus (Nelumbo nucifera Gaertn.). Genome Biol. 2013, 14, R41. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Fan, G.; Liu, Y.; Sun, F.; Shi, C.; Liu, X.; Peng, J.; Chen, W.; Huang, X.; Cheng, S.; et al. The sacred lotus genome provides insights into the evolution of flowering plants. Plant J. 2013, 76, 557–567. [Google Scholar] [CrossRef] [PubMed]
- Gandolfo, M.A.; Cuneo, R.N. Fossil Nelumbonaceae from the La Colonia Formation (Campanian-Maastrichtian, Upper Cretaceous), Chubut, Patagonia, Argentina. Rev. Palaeobot. Palynol. 2005, 133, 169–178. [Google Scholar] [CrossRef]
- Cronquist, A. An Integrated System of Classification of Flowering Plants; Columbia University Press: New York, NY, USA, 1981. [Google Scholar]
- Dahlgren, G. An updated angiosperm classification. Bot. J. Linn. Soc. 1989, 100, 197–203. [Google Scholar] [CrossRef]
- Thorne, R.F. An Updated Phylogenetic Classification of the Flowering Plants. Aliso 1992, 13, 365–389. [Google Scholar] [CrossRef]
- Takhtajan, A. Diversity and Classification of Flowering Plants; Columbia University Press: New York, NY, USA, 1997. [Google Scholar]
- Stevens, P.F. (2001 onwards). Angiosperm Phylogeny Website, Version 14; July 2017. Available online: http://www.mobot.org/MOBOT/research/APweb/ (accessed on 23 June 2019).
- Byng, J.; Chase, M.; Christenhusz, M.; Fay, M.; Judd, W.; Mabberley, D.; Sennikov, A.; Soltis, D.; Soltis, P.; Stevens, P. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar]
- Yoo, M.J.; Soltis, P.S.; Soltis, D.E. Expression of floral MADS-box genes in two divergent water lilies: Nymphaeales and Nelumbo. Int. J. Plant Sci. 2010, 171, 121–146. [Google Scholar] [CrossRef]
- Diao, Y.; Chen, L.; Yang, G.; Zhou, M.; Song, Y. Nuclear DNA C-values in 12 species in Nymphaeales. Caryologia 2006, 59, 25–30. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Nyong, A.T.; Shi, T.; Yang, P. The complexity of alternative splicing and landscape of tissue-specific expression in lotus (Nelumbo nucifera) unveiled by Illumina- and single-molecule real-time-based RNA-sequencing. DNA Res. 2019. [Google Scholar] [CrossRef] [PubMed]
- Gui, S.; Peng, J.; Wang, X.; Wu, Z.; Cao, R.; Salse, J.; Zhang, H.; Zhu, Z.; Xia, Q.; Quan, Z.; et al. Improving Nelumbo nucifera genome assemblies using high-resolution genetic maps and BioNano genome mapping reveals ancient chromosome rearrangements. Plant J. 2018, 94, 721–734. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Deng, J.; Damaris, R.N.; Yang, M.; Xu, L.; Yang, P. LOTUS-DB: An integrative and interactive database for Nelumbo nucifera study. Database 2015, 2015, bav023. [Google Scholar] [CrossRef] [PubMed]
- Gui, S.; Wu, Z.; Zhang, H.; Zheng, Y.; Zhu, Z.; Liang, D.; Ding, Y. The mitochondrial genome map of Nelumbo nucifera reveals ancient evolutionary features. Sci. Rep. 2016, 6, 30158. [Google Scholar] [CrossRef]
- Wu, Z.; Gui, S.; Quan, Z.; Pan, L.; Wang, S.; Ke, W.; Liang, D.; Ding, Y. A precise chloroplast genome of Nelumbo nucifera (Nelumbonaceae) evaluated with Sanger, Illumina MiSeq, and PacBio RS II sequencing platforms: Insight into the plastid evolution of basal eudicots. BMC Plant Biol. 2014, 14, 289. [Google Scholar] [CrossRef]
- Shi, T.; Wang, K.; Yang, P. The evolution of plant microRNAs: Insights from a basal eudicot sacred lotus. Plant J. 2017, 89, 442–457. [Google Scholar] [CrossRef] [PubMed]
- Meng, Z.; Hu, X.; Zhang, Z.; Li, Z.; Lin, Q.; Yang, M.; Yang, P.; Ming, R.; Yu, Q.; Wang, K. Chromosome Nomenclature and Cytological Characterization of Sacred Lotus. Cytogenet. Genome Res. 2017, 153, 223–231. [Google Scholar] [CrossRef]
- Shen-Miller, J.; Mudgett, M.B.; Schopf, J.W.; Clarke, S.; Berger, R. Exceptional seed longevity and robust growth: Ancient Sacred Lotus from China. Am. J. Bot. 1995, 82, 1367–1380. [Google Scholar] [CrossRef]
- Van Bergen, P.F.; Hatcher, P.G.; Boon, J.J.; Collinson, M.E.; de Leeuw, J.W. Macromolecular composition of the propagule wall of Nelumbo nucifera. Phytochemistry 1997, 45, 601–610. [Google Scholar] [CrossRef]
- Liu, Y.; Ma, S.S.; Ibrahim, S.A.; Li, E.H.; Yang, H.; Huang, W. Identification and antioxidant properties of polyphenols in lotus seed epicarp at different ripening stages. Food Chem. 2015, 185, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Shen-Miller, J.; Lindner, P.; Xie, Y.; Villa, S.; Wooding, K.; Clarke, S.G.; Loo, R.R.; Loo, J.A. Thermal-stable proteins of fruit of long-living Sacred Lotus Nelumbo nucifera Gaertn var. China Antique. Trop. Plant Biol. 2013, 6, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Chu, P.; Chen, H.; Zhou, Y.; Li, Y.; Ding, Y.; Jiang, L.; Tsang, E.W.; Wu, K.; Huang, S. Proteomic and functional analyses of Nelumbo nucifera annexins involved in seed thermotolerance and germination vigor. Planta 2012, 235, 1271–1288. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.H.; Chu, P.; Zhou, Y.L.; Ding, Y.; Li, Y.; Liu, J.; Jiang, L.W.; Huang, S.Z. Ectopic expression of NnPER1, a Nelumbo nucifera 1-cysteine peroxiredoxin antioxidant, enhances seed longevity and stress tolerance in Arabidopsis. Plant J. 2016, 88, 608–619. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Jin, J.; Qian, Q.; Huang, K.; Ding, Y. Small RNA and degradome profiling reveals miRNA regulation in the seed germination of ancient eudicot Nelumbo nucifera. BMC Genom. 2016, 17, 684. [Google Scholar] [CrossRef]
- Darmanin, T.; Guittard, F. Superhydrophobic and superoleophobic properties in nature. Mater. Today 2015, 18, 273–285. [Google Scholar] [CrossRef]
- Ensikat, H.J.; Ditsche-Kuru, P.; Neinhuis, C.; Barthlott, W. Superhydrophobicity in perfection: The outstanding properties of the lotus leaf. Beilstein J. Nanotechnol. 2011, 2, 152–161. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, H.; Yu, X.; Chen, F.; Wu, J. Microscopic Observations of the Lotus Leaf for Explaining the Outstanding Mechanical Properties. J. Bionic Eng. 2012, 9, 84–90. [Google Scholar] [CrossRef]
- Marmur, A. The Lotus effect: Superhydrophobicity and metastability. Langmuir 2004, 20, 3517–3519. [Google Scholar] [CrossRef]
- Bhushan, B.; Jung, Y.C.; Koch, K. Self-Cleaning Efficiency of Artificial Superhydrophobic Surfaces. Langmuir 2009, 25, 3240–3248. [Google Scholar] [CrossRef]
- Yang, X.; Wang, Z.; Feng, T.; Li, J.; Huang, L.; Yang, B.; Zhao, H.; Jenks, M.A.; Yang, P.; Lü, S. Evolutionarily conserved function of the sacred lotus (Nelumbo nucifera Gaertn.) CER2-LIKE family in very-long-chain fatty acid elongation. Planta 2018, 248, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Li, J.K.; Huang, S.Q. Flower thermoregulation facilitates fertilization in Asian sacred lotus. Ann. Bot. 2009, 103, 1159–1163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watling, J.; Robinson, S.; Seymour, R. Contribution of the Alternative Pathway to Respiration during Thermogenesis in Flowers of the Sacred Lotus. Plant Physiol. 2006, 140, 1367–1373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seymour, R.; Schultze-Motel, P.; Lamprecht, I. Heat production by sacred lotus flowers depends on ambient temperature, not light cycle. J. Exp. Bot. 1998, 49, 1213–1217. [Google Scholar] [CrossRef]
- Grant, N.; Miller, R.; Watling, J.; Robinson, S. Distribution of thermogenic activity in floral tissues of Nelumbo nucifera. Funct. Plant Biol. 2010, 37, 1085–1095. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Lu, J.; Wang, J.; Chen, F.; Leng, F.; Li, H. Regulation of thermogenesis in plants: The interaction of alternative oxidase and plant uncoupling mitochondrial protein. J. Integr. Plant Biol. 2011, 53, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhang, Z. Floral thermogenesis: An adaptive strategy of pollination biology in Magnoliaceae. Commun. Integr. Biol. 2015, 8, e992746. [Google Scholar] [CrossRef]
- Wagner, A.M.; Krab, K.; Wagner, M.J.; Moore, A.L. Regulation of thermogenesis in flowering Araceae: The role of the alternative oxidase. Biochim. Biophys. Acta 2008, 1777, 993–1000. [Google Scholar] [CrossRef] [Green Version]
- Dieringer, G.; Leticia Cabrera, R.; Mottaleb, M. Ecological relationship between floral thermogenesis and pollination in Nelumbo lutea (Nelumbonaceae). Am. J. Bot. 2014, 101, 357–364. [Google Scholar] [CrossRef] [Green Version]
- Miller, R.E.; Watling, J.R.; Robinson, S.A. Functional transition in the floral receptacle of the sacred lotus (Nelumbo nucifera): From thermogenesis to photosynthesis. Funct. Plant Biol. 2009, 36, 471–480. [Google Scholar] [CrossRef]
- Grant, N.M.; Miller, R.E.; Watling, J.R.; Robinson, S.A. Synchronicity of thermogenic activity, alternative pathway respiratory flux, AOX protein content, and carbohydrates in receptacle tissues of sacred lotus during floral development. J. Exp. Bot. 2008, 59, 705–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Liu, X.Q.; Gituru, R.W.; Juntawong, N.; Zhou, M.Q.; Chen, L.Q. Genetic diversity and classification of Nelumbo germplasm of different origins by RAPD and ISSR analysis. Sci. Hortic. 2010, 125, 724–732. [Google Scholar] [CrossRef]
- Hu, J.; Pan, L.; Liu, H.; Wang, S.; Wu, Z.; Ke, W.; Ding, Y. Comparative analysis of genetic diversity in sacred lotus (Nelumbo nucifera Gaertn.) using AFLP and SSR markers. Mol. Biol. Rep. 2012, 39, 3637–3647. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Han, Y.; VanBuren, R.; Ming, R.; Xu, L.; Han, Y.; Liu, Y. Genetic linkage maps for Asian and American lotus constructed using novel SSR markers derived from the genome of sequenced cultivar. BMC Genom. 2012, 13, 653. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Han, Y.N.; Xu, L.M.; Zhao, J.R.; Liu, Y.L. Comparative analysis of genetic diversity of lotus (Nelumbo) using SSR and SRAP markers. Sci. Hortic. 2012, 142, 185–195. [Google Scholar] [CrossRef]
- Huang, L.; Yang, M.; Li, L.; Li, H.; Yang, D.; Shi, T.; Yang, P. Whole genome re-sequencing reveals evolutionary patterns of sacred lotus (Nelumbo nucifera). J. Integr. Plant Biol. 2018, 60, 2–15. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Yang, J.-X.; Mao, T.-Y.; Zhu, H.-H.; Xiang, L.; Zhang, J.; Chen, L.-Q. Detection of Highly Differentiated Genomic Regions Between Lotus (Nelumbo nucifera Gaertn.) with Contrasting Plant Architecture and Their Functional Relevance to Plant Architecture. Front. Plant Sci. 2018, 9, 1219. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Gui, S.; Zhu, Z.; Wang, X.; Ke, W.; Ding, Y. Genome-Wide Identification of SSR and SNP Markers Based on Whole-Genome Re-Sequencing of a Thailand Wild Sacred Lotus (Nelumbo nucifera). PLoS ONE 2015, 10, e0143765. [Google Scholar] [CrossRef]
- Deng, J.; Chen, S.; Yin, X.J.; Wang, K.; Liu, Y.L.; Li, S.H.; Yang, P.F. Systematic qualitative and quantitative assessment of anthocyanins, flavones and flavonols in the petals of 108 lotus (Nelumbo nucifera) cultivars. Food Chem. 2013, 139, 307–312. [Google Scholar] [CrossRef]
- Deng, J.; Li, M.; Huang, L.; Yang, M.; Yang, P. Genome-wide analysis of the R2R3 MYB subfamily genes in lotus (Nelumbo nucifera). Plant Mol. Biol. Report. 2016, 34, 1016–1026. [Google Scholar] [CrossRef]
- Sun, S.-S.; Gugger, P.F.; Wang, Q.-F.; Chen, J.-M. Identification of a R2R3-MYB gene regulating anthocyanin biosynthesis and relationships between its variation and flower color difference in lotus (Nelumbo Adans.). PeerJ 2016, 4, e2369. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Fu, Z.; Chen, S.; Damaris, R.N.; Wang, K.; Li, T.; Yang, P. Proteomic and Epigenetic Analyses of Lotus (Nelumbo nucifera) Petals Between Red and White cultivars. Plant Cell Physiol. 2015, 56, 1546. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Damaris, R.N.; Shi, T.; Li, J.; Yang, P. Transcriptomic analysis identifies the key genes involved in stamen petaloid in lotus (Nelumbo nucifera). BMC Genom. 2018, 19, 554. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Liu, M.; Damaris, R.N.; Nyong’a, T.M.; Cao, D.; Ou, K.; Yang, P. Genome-Wide DNA Methylation Profiling in the Lotus (Nelumbo nucifera) Flower Showing its Contribution to the Stamen Petaloid. Plants 2019, 8, 135. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Zhu, L.; Xu, L.; Pan, C.; Liu, Y. Comparative transcriptomic analysis of the regulation of flowering in temperate and tropical lotus (Nelumbo nucifera) by RNA-Seq. Ann. Appl. Biol. 2014, 165, 73–95. [Google Scholar] [CrossRef]
- Yang, M.; Zhu, L.; Pan, C.; Xu, L.; Liu, Y.; Ke, W.; Yang, P. Transcriptomic Analysis of the Regulation of Rhizome Formation in Temperate and Tropical Lotus (Nelumbo nucifera). Sci. Rep. 2015, 5, 13059. [Google Scholar] [CrossRef]
- Kim, M.-J.; Nelson, W.; Soderlund, C.A.; Gang, D.R. Next-Generation Sequencing-Based Transcriptional Profiling of Sacred Lotus ‘China Antique’. Trop. Plant Biol. 2013, 6, 161–179. [Google Scholar] [CrossRef]
- Yang, M.; Xu, L.; Liu, Y.; Yang, P. RNA-Seq Uncovers SNPs and Alternative Splicing Events in Asian Lotus (Nelumbo nucifera). PLoS ONE 2015, 10, e0125702. [Google Scholar] [CrossRef]
- Li, J.; Shi, T.; Huang, L.; He, D.; Nyong’A, T.M.; Yang, P. Systematic transcriptomic analysis provides insights into lotus (Nelumbo nucifera) seed development. Plant Growth Regul. 2018, 86, 339–350. [Google Scholar] [CrossRef]
- Chen, S.; Fang, L.; Xi, H.; Guan, L.; Fang, J.; Liu, Y.; Wu, B.; Li, S. Simultaneous qualitative assessment and quantitative analysis of flavonoids in various tissues of lotus (Nelumbo nucifera) using high performance liquid chromatography coupled with triple quad mass spectrometry. Anal. Chim. Acta 2012, 724, 127–135. [Google Scholar] [CrossRef]
- Limwachiranon, J.; Huang, H.; Shi, Z.; Li, L.; Luo, Z. Lotus Flavonoids and Phenolic Acids: Health Promotion and Safe Consumption Dosages. Compr. Rev. Food Sci. Food Saf. 2018, 17, 458–471. [Google Scholar] [CrossRef] [Green Version]
- Sharma, B.R.; Gautam, L.N.; Adhikari, D.; Karki, R. A Comprehensive Review on Chemical Profiling of Nelumbo Nucifera: Potential for Drug Development. Phytother. Res. 2017, 31, 3–26. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Xiang, Y.; Deng, J.; Liu, Y.L.; Li, S.H. Simultaneous Analysis of Anthocyanin and Non-Anthocyanin Flavonoid in Various Tissues of Different Lotus (Nelumbo) Cultivars by HPLC-DAD-ESI-MSn. PLoS ONE 2013, 8, e62291. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zheng, Y.; Fang, J.B.; Liu, Y.L.; Li, S.H. Flavonoids in lotus (Nelumbo) leaves evaluated by HPLC-MSn at the germplasm level. Food Res. Int. 2013, 54, 796–803. [Google Scholar] [CrossRef]
- Li, S.S.; Wu, J.; Chen, L.G.; Du, H.; Xu, Y.J.; Wang, L.J.; Zhang, H.J.; Zheng, X.C.; Wang, L.S. Biogenesis of C-Glycosyl Flavones and Profiling of Flavonoid Glycosides in Lotus (Nelumbo nucifera). PLoS ONE 2014, 9, e108860. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Shen, J.; Chang, K.J.; Kim, S.H. Comparative Analysis of Antioxidant Activity and Functional Components of the Ethanol Extract of Lotus (Nelumbo nucifera) from Various Growing Regions. J. Agric. Food Chem. 2014, 62, 6227–6235. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.Z.; Wu, W.; Jiao, L.L.; Yang, P.F.; Guo, M.Q. Analysis of Flavonoids in Lotus (Nelumbo nucifera) Leaves and Their Antioxidant Activity Using Macroporous Resin Chromatography Coupled with LC-MS/MS and Antioxidant Biochemical Assays. Molecules 2015, 20, 10553–10565. [Google Scholar] [CrossRef]
- Feng, C.Y.; Li, S.S.; Yin, D.D.; Zhang, H.J.; Tian, D.K.; Wu, Q.; Wang, L.J.; Su, S.; Wang, L.S. Rapid determination of flavonoids in plumules of sacred lotus cultivars and assessment of their antioxidant activities. Ind. Crop. Prod. 2016, 87, 96–104. [Google Scholar] [CrossRef]
- Guo, Y.J.; Chen, X.; Qi, J.; Yu, B.Y. Simultaneous qualitative and quantitative analysis of flavonoids and alkaloids from the leaves of Nelumbo nucifera Gaertn. using high-performance liquid chromatography with quadrupole time-of-flight mass spectrometry. J. Sep. Sci. 2016, 39, 2499–2507. [Google Scholar] [CrossRef]
- Zhu, M.Z.; Liu, T.; Zhang, C.Y.; Guo, M.Q. Flavonoids of Lotus (Nelumbo nucifera) Seed Embryos and Their Antioxidant Potential. J. Food Sci. 2017, 82, 1834–1841. [Google Scholar] [CrossRef]
- Tian, W.Y.; Zhi, H.; Yang, C.; Wang, L.K.; Long, J.T.; Xiao, L.M.; Liang, J.Z.; Huang, Y.; Zheng, X.; Zhao, S.Q.; et al. Chemical composition of alkaloids of Plumula nelumbinis and their antioxidant activity from different habitats in China. Ind. Crop. Prod. 2018, 125, 537–548. [Google Scholar] [CrossRef]
- Zheng, J.X.; Tian, W.Y.; Yang, C.; Shi, W.P.; Cao, P.H.; Long, J.T.; Xiao, L.M.; Wu, Y.; Liang, J.Z.; Li, X.B.; et al. Identification of flavonoids in Plumula nelumbinis and evaluation of their antioxidant properties from different habitats. Ind. Crop. Prod. 2019, 127, 36–45. [Google Scholar] [CrossRef]
- Commission, C.P. Pharmacopoeia of the People’s Republic of China; China Medical Science and Technology Press: Beijing, China, 2015. [Google Scholar]
- Huang, B.; Ban, X.; He, J.; Tong, J.; Tian, J.; Wang, Y. Hepatoprotective and antioxidant activity of ethanolic extracts of edible lotus (Nelumbo nucifera Gaertn.) leaves. Food Chem. 2010, 120, 873–878. [Google Scholar] [CrossRef]
- Paudel, K.R.; Panth, N. Phytochemical Profile and Biological Activity of Nelumbo nucifera. Evid. -Based Complement. Altern. Med. 2015, 2015, 789124. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.H.; He, X.X.; Kong, L.T.; Liao, Y.H.; Pan, R.L.; Xiao, B.X.; Liu, X.M.; Chang, Q. Identification and characterization of potent CYP2D6 inhibitors in lotus leaves. J. Ethnopharmacol. 2014, 153, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.-H.; He, X.-X.; You, C.; Tao, X.; Wang, L.-S.; Zhang, M.-D.; Zhou, Y.-F.; Chang, Q. Pharmacokinetics of Nuciferine and N-Nornuciferine, Two Major Alkaloids From Nelumbo nucifera Leaves, in Rat Plasma and the Brain. Front. Pharmacol. 2018, 9, 902. [Google Scholar] [CrossRef]
- Yang, M.; Zhu, L.; Li, L.; Li, J.; Xu, L.; Feng, J.; Liu, Y. Digital Gene Expression Analysis Provides Insight into the Transcript Profile of the Genes Involved in Aporphine Alkaloid Biosynthesis in Lotus (Nelumbo nucifera). Front. Plant Sci. 2017, 8, 80. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Zhao, L.; Fang, T.; Xiong, Y.; Ogutu, C.; Yang, D.; Vimolmangkang, S.; Liu, Y.; Han, Y. Investigation of benzylisoquinoline alkaloid biosynthetic pathway and its transcriptional regulation in lotus. Hortic. Res. 2018, 5, 29. [Google Scholar] [CrossRef]
- Menendez-Perdomo, I.M.; Facchini, P.J. Benzylisoquinoline Alkaloids Biosynthesis in Sacred Lotus. Molecules 2018, 23, 2899. [Google Scholar] [CrossRef]
- Arunyanart, S.; Chaitrayagun, M. Induction of somatic embryogenesis in lotus (Nelumbo nucifera Geartn.). Sci. Hortic. 2005, 105, 411–420. [Google Scholar] [CrossRef]
- Liu, Y.; Chaturvedi, P.; Fu, J.; Cai, Q.; Weckwerth, W.; Yang, P. Induction and quantitative proteomic analysis of cell dedifferentiation during callus formation of lotus (Nelumbo nucifera Gaertn. spp. baijianlian). J. Proteom. 2016, 131, 61–70. [Google Scholar] [CrossRef]
- Shou, S.Y.; Miao, L.X.; Zai, W.S.; Huang, X.Z.; Guo, D.P. Factors influencing shoot multiplication of lotus (Nelumbo nucifera). Biol. Plant. 2008, 52, 529–532. [Google Scholar] [CrossRef]
- Buathong, R.; Saetiew, K.; Phansiri, S.; Parinthawong, N.; Arunyanart, S. Tissue culture and transformation of the antisense DFR gene into lotus (Nelumbo nucifera Gaertn.) through particle bombardment. Sci. Hortic. 2013, 161, 216–222. [Google Scholar] [CrossRef]
- Kanjana, S.; Prissadang, A.; Nanglak, P.; Sumay, A. Transformation of Antisense Chalcone Synthase (CHS) Gene into Lotus (Nelumbo Nucifera Gaertn.) by Particle Bombardment. Open Biotechnol. J. 2017, 11, 1–8. [Google Scholar] [CrossRef]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, Z.; Zhang, C.; Cao, D.; Damaris, R.N.; Yang, P. The Latest Studies on Lotus (Nelumbo nucifera)-an Emerging Horticultural Model Plant. Int. J. Mol. Sci. 2019, 20, 3680. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20153680
Lin Z, Zhang C, Cao D, Damaris RN, Yang P. The Latest Studies on Lotus (Nelumbo nucifera)-an Emerging Horticultural Model Plant. International Journal of Molecular Sciences. 2019; 20(15):3680. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20153680
Chicago/Turabian StyleLin, Zhongyuan, Cheng Zhang, Dingding Cao, Rebecca Njeri Damaris, and Pingfang Yang. 2019. "The Latest Studies on Lotus (Nelumbo nucifera)-an Emerging Horticultural Model Plant" International Journal of Molecular Sciences 20, no. 15: 3680. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20153680